6IOV
 
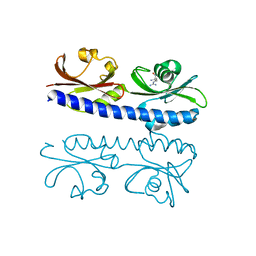 | | The ligand binding domain of Mlp37 with arginine | | Descriptor: | ARGININE, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis of the binding affinity of chemoreceptors Mlp24p and Mlp37p for various amino acids.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
6IOQ
 
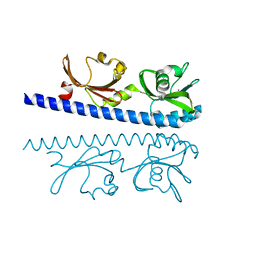 | | The ligand binding domain of Mlp24 with glycine | | Descriptor: | CALCIUM ION, GLYCINE, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOU
 
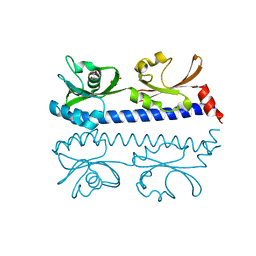 | | The ligand binding domain of Mlp24 with serine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOR
 
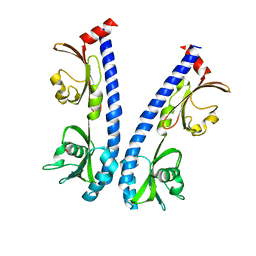 | | The ligand binding domain of Mlp24 with asparagine | | Descriptor: | ASPARAGINE, CALCIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOT
 
 | | The ligand binding domain of Mlp24 with arginine | | Descriptor: | ARGININE, CALCIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOS
 
 | | The ligand binding domain of Mlp24 with proline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
5ETY
 
 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | Descriptor: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | Authors: | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
5AVE
 
 | | The ligand binding domain of Mlp37 with serine | | Descriptor: | Methyl-accepting chemotaxis (MCP) signaling domain protein, SERINE | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
5AVF
 
 | | The ligand binding domain of Mlp37 with taurine | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
6IOP
 
 | | The ligand binding domain of Mlp24 | | Descriptor: | ACETATE ION, ALANINE, CALCIUM ION, ... | | Authors: | Sumita, K, Takahashi, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
7CE4
 
 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
6AD9
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-9 | | Descriptor: | 12-mer peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, 3-[(1E)-1-{8-[(4-methyl-2-propyl-1H-benzimidazol-1-yl)methyl]dibenzo[b,e]oxepin-11(6H)-ylidene}ethyl]-1,2,4-oxadiazol-5(4H)-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Takahashi, Y, Suzuki, M, Yamamoto, K, Saito, J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Dihydrodibenzooxepine Peroxisome Proliferator-Activated Receptor (PPAR) Gamma Ligands of a Novel Binding Mode as Anticancer Agents: Effective Mimicry of Chiral Structures by Olefinic E/ Z-Isomers.
J. Med. Chem., 61, 2018
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
5WT2
 
 | | NifS from Helicobacter pylori | | Descriptor: | CHLORIDE ION, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL, ... | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 2019
|
|
5WT6
 
 | |
5WT5
 
 | | L-homocysteine-bound NifS from Helicobacter pylori | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural snapshot of cysteine desulfurase NifS with L-cysteine in initiation of catalysis
to be published
|
|
1WTF
 
 | | Crystal structure of Bacillus thermoproteolyticus Ferredoxin Variants Containing Unexpected [3Fe-4S] Cluster that is linked to Coenzyme A at 1.6 A Resolution | | Descriptor: | COENZYME A, FE3-S4 CLUSTER, Ferredoxin, ... | | Authors: | Shirakawa, T, Takahashi, Y, Wada, K, Hirota, J, Takao, T, Ohmori, D, Fukuyama, K. | | Deposit date: | 2004-11-22 | | Release date: | 2005-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of variant molecules of Bacillus thermoproteolyticus ferredoxin: crystal structure reveals bound coenzyme A and an unexpected [3Fe-4S] cluster associated with a canonical [4Fe-4S] ligand motif
Biochemistry, 44, 2005
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
1IQZ
 
 | | OXIDIZED [4Fe-4S] FERREDOXIN FROM BACILLUS THERMOPROTEOLYTICUS (FORM I) | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Fukuyama, K, Okada, T, Kakuta, Y, Takahashi, Y. | | Deposit date: | 2001-08-30 | | Release date: | 2002-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: systematic distortion of [4Fe-4S] cluster in the protein.
J.Mol.Biol., 315, 2002
|
|
1IR0
 
 | | OXIDIZED [4Fe-4S] FERREDOXIN FROM BACILLUS THERMOPROTEOLYTICUS (FORM II) | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Fukuyama, K, Okada, T, Kakuta, Y, Takahashi, Y. | | Deposit date: | 2001-08-30 | | Release date: | 2002-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: systematic distortion of [4Fe-4S] cluster in the protein.
J.Mol.Biol., 315, 2002
|
|
5H41
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose, isofagomine, sulfate ion | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, SULFATE ION, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H40
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H3Z
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H42
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with alpha-d-glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Uncharacterized protein, alpha-D-glucopyranose | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
