3WVS
 
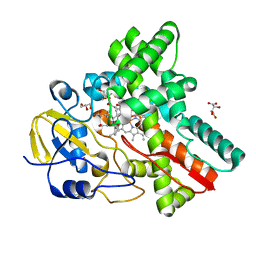 | | Crystal Structure of Cytochrome P450revI | | Descriptor: | (2E,4S,5S,6E,8E)-10-{(2R,3S,6S,8R,9S)-9-butyl-8-[(1E,3E)-4-carboxy-3-methylbuta-1,3-dien-1-yl]-3-methyl-1,7-dioxaspiro[5.5]undec-2-yl}-5-hydroxy-4,8-dimethyldeca-2,6,8-trienoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Nagano, S, Takahashi, S, Osada, H, Shiro, Y. | | Deposit date: | 2014-06-06 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses of cytochrome P450revI involved in reveromycin A biosynthesis and evaluation of the biological activity of its substrate, reveromycin T.
J.Biol.Chem., 289, 2014
|
|
8KGA
 
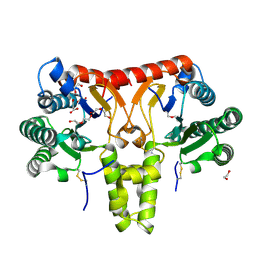 | | SlNDPS1-AtcPT4 Chimera | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimethylallylcistransferase CPT1, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Biosynthesising various rubber-like polymers by reconstituting prenyltransferases on Hevea rubber particles: molecular and structural bases of the versatile system
To Be Published
|
|
8KGB
 
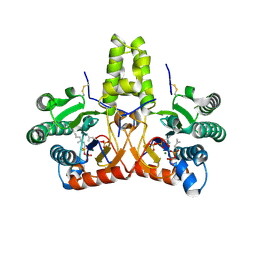 | | SlNDPS1-AtcPT4 Chimera complexed with GSPP, Mg2+, and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Dimethylallylcistransferase CPT1, chloroplastic,Dehydrodolichyl diphosphate synthase 2, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A versatile system for enzymatic synthesis of natural and unnatural polyisoprenoids on rubber particles
To Be Published
|
|
7BUR
 
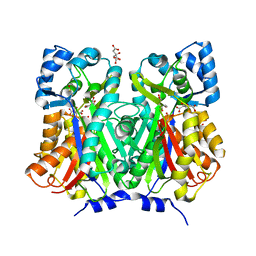 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin | | Descriptor: | CITRIC ACID, Chalcone synthase 1, NARINGENIN | | Authors: | Imaizumi, R, Mameda, R, Takeshita, K, Waki, T, Kubo, H, Sakai, N, Nakata, S, Takahashi, S, Kataoka, K, Yamamoto, M, Yamashita, S, Nakayama, T. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean.
Proteins, 2020
|
|
7BUS
 
 | | Chalcone synthase from Glycine max (L.) Merr (soybean) | | Descriptor: | Chalcone synthase | | Authors: | Imaizumi, R, Mameda, R, Takeshita, K, Waki, T, Kubo, H, Sakai, N, Nakata, S, Takahashi, S, Kataoka, K, Yamamoto, M, Yamashita, S, Nakayama, T. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean.
Proteins, 2020
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
1IQ0
 
 | | THERMUS THERMOPHILUS ARGINYL-TRNA SYNTHETASE | | Descriptor: | ARGINYL-TRNA SYNTHETASE | | Authors: | Shimada, A, Nureki, O, Goto, M, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational studies of the recognition of the arginine tRNA-specific major identity element, A20, by arginyl-tRNA synthetase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1ERZ
 
 | | CRYSTAL STRUCTURE OF N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE WITH A NOVEL CATALYTIC FRAMEWORK COMMON TO AMIDOHYDROLASES | | Descriptor: | N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE | | Authors: | Nakai, T, Hasegawa, T, Yamashita, E, Yamamoto, M, Kumasaka, T, Ueki, T, Nanba, H, Ikenaka, Y, Takahashi, S, Sato, M, Tsukihara, T. | | Deposit date: | 2000-04-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases.
Structure Fold.Des., 8, 2000
|
|
5ING
 
 | | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender unit | | Descriptor: | Putative carboxyl transferase | | Authors: | Valentic, T.R, Ray, L, Miyazawa, T, Song, L, Withall, D.M, Milligan, J.C, Takahashi, S, Osada, H, Tsai, S.C, Challis, G.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units.
Nat Commun, 7, 2016
|
|
7VPC
 
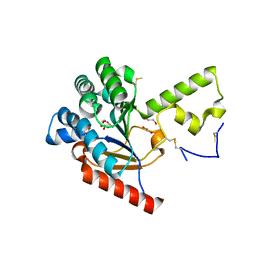 | | Neryl diphosphate synthase from Solanum lycopersicum | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Misawa, S, Takeshita, K, Sakai, N, Yamamoto, M, Kataoka, K, Nakayama, T, Takahashi, S, Yamashita, S. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-based engineering of a short-chain cis-prenyltransferase to biosynthesize nonnatural all-cis-polyisoprenoids: molecular mechanisms for primer substrate recognition and ultimate product chain-length determination.
Febs J., 289, 2022
|
|
8X35
 
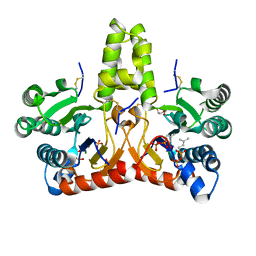 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X37
 
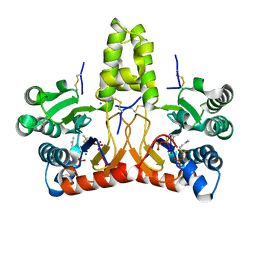 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X36
 
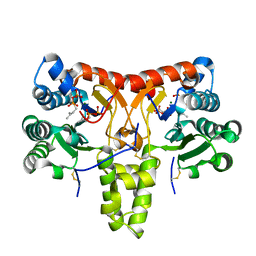 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form B) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
6GIW
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum (Mutation L91P) with Chlorophyll-a | | Descriptor: | CHLOROPHYLL A, Water-soluble chlorophyll protein | | Authors: | Palm, D.M, Agostini, A, Averesch, V, Girr, P, Werwie, M, Takahashi, S, Satoh, H, Jaenicke, E, Paulsen, H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein.
Nat Plants, 4, 2018
|
|
6GIX
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum (Mutation L91P) with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-soluble chlorophyll protein | | Authors: | Palm, D.M, Agostini, A, Averesch, V, Girr, P, Werwie, M, Takahashi, S, Satoh, H, Jaenicke, E, Paulsen, H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein.
Nat Plants, 4, 2018
|
|
1UF8
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-Phenylalanine | | Descriptor: | D-[(AMINO)CARBONYL]PHENYLALANINE, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF4
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase | | Descriptor: | N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid
To be published
|
|
1UF7
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-valine | | Descriptor: | 3-METHYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF5
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-methionine | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLSULFANYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
3VHX
 
 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
2E89
 
 | | Crystal structure of Aquifex aeolicus TilS in a complex with ATP, Magnesium ion, and L-lysine | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LYSINE, MAGNESIUM ION, ... | | Authors: | Kuratani, M, Yoshikawa, Y, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine
To be Published
|
|
7E5V
 
 | | Crystal structure of Phm7 in complex with inhibitor | | Descriptor: | Diels-Alderase, GLYCEROL, SULFATE ION, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5U
 
 | | Crystal structure of Phm7 | | Descriptor: | CHLORIDE ION, Diels-Alderase, GLYCEROL, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5T
 
 | | Crystal structure of Fsa2 | | Descriptor: | Diels-Alderase fsa2, ETHANOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16977525 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CT4
 
 | | Crystal structure of D-amino acid oxidase from Rasamsonia emersonii strain YA | | Descriptor: | D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Shimekake, Y, Hirato, Y, Okazaki, S, Funabashi, R, Goto, M, Furuichi, T, Suzuki, H, Takahashi, S. | | Deposit date: | 2020-08-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of a unique D-amino-acid oxidase from the thermophilic fungus Rasamsonia emersonii strain YA.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
