7SA2
 
 | |
7SIS
 
 | |
7U1R
 
 | |
7RTD
 
 | |
7RTR
 
 | |
7UR1
 
 | |
7UM2
 
 | |
7N4K
 
 | | 6218 TCR in complex with H2-Db PA 224 | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-04 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5C
 
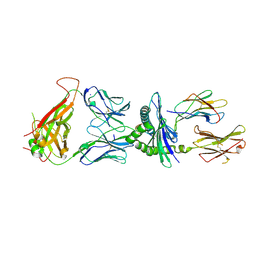 | | 6218 TCR in complex with H2Db PA with an engineered TCR-pMHC disulfide bond | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 with T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-05 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5P
 
 | | 6218 TCR in complex with H2-Db PA224-233 with a cysteine mutant | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5Q
 
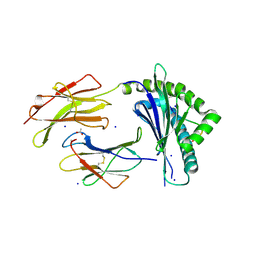 | |
7KGS
 
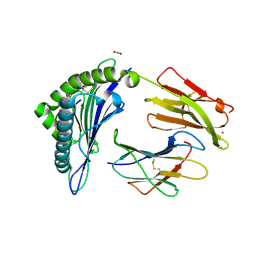 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N138-146 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
7KGT
 
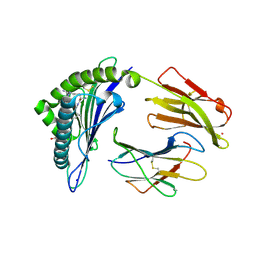 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N226-234 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
7KGR
 
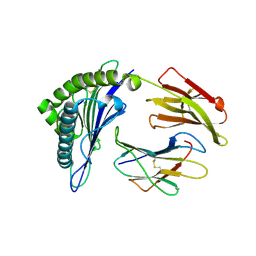 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N159-167 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Nucleoprotein | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGQ
 
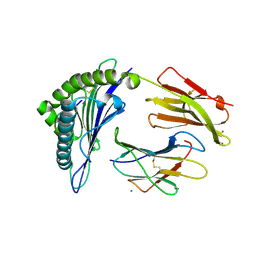 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N222-230 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGP
 
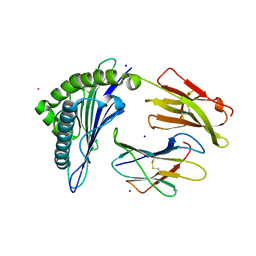 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N316-324 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGO
 
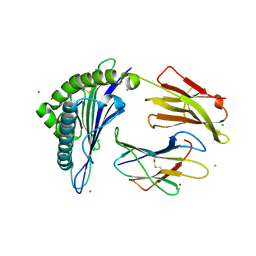 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N351-359 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
6XQA
 
 | | Crystal Structure of HLA A*2402 in complex with TYQWVLKNL, an 9-mer epitope from Influenza B virus | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
7TLT
 
 | | SARS-CoV-2 Spike-derived peptide S489-497 (YFPLQSYGF) presented by HLA-A*29:02 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Murdolo, L.D, Szeto, C, Gras, S. | | Deposit date: | 2022-01-18 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ablation of CD8 + T cell recognition of an immunodominant epitope in SARS-CoV-2 Omicron variants BA.1, BA.2 and BA.3.
Nat Commun, 13, 2022
|
|
7UC5
 
 | |
8RBV
 
 | | SARS-CoV-2 Spike-derived peptide S976-984 S982A mutant (VLNDILARL) presented by HLA-A*02:01 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8REF
 
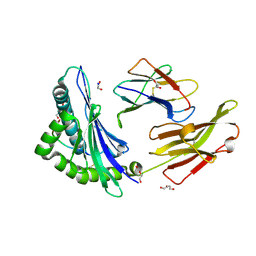 | | Crystal structure of HLA B*13:01 in complex with SVLNDILARL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RH6
 
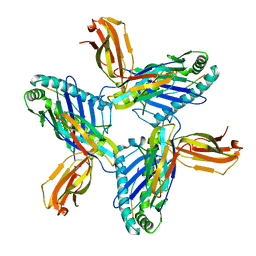 | | Crystal structure of HLA-A*11:01 in complex with SVLNDILSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RBU
 
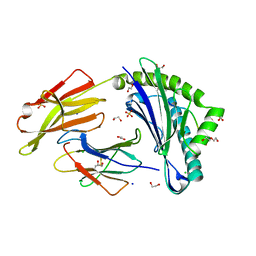 | | Crystal structure of HLA-A*11:01 in complex with SVLNDILARL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RCV
 
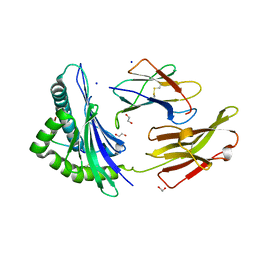 | | Crystal structure of HLA B*13:01 in complex with SVLNDIFSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen B alpha chain, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
