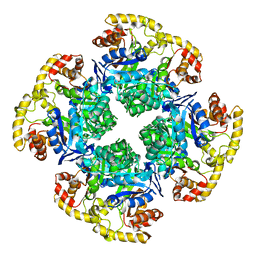
6CI1
 
 | |
4V47
 
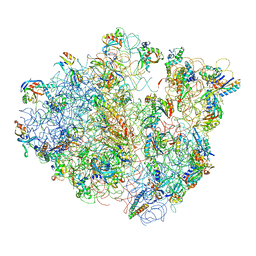 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
8VT0
 
 | | SPOT-RASTR - a cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Esfahani, B.G, Randolph, P, Peng, R, Grant, T, Stroupe, M.E, Stagg, S.M. | | Deposit date: | 2024-01-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | SPOT-RASTR-A cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface.
Pnas Nexus, 3, 2024
|
|
6OM6
 
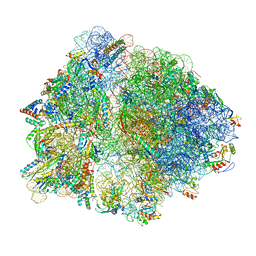 | | Structure of trans-translation inhibitor bound to E. coli 70S ribosome with P site tRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Mehrani, A, Keiler, K.C, Stagg, S.M, Dunham, C.M. | | Deposit date: | 2019-04-18 | | Release date: | 2021-02-10 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | trans-Translation inhibitors bind to a novel site on the ribosome and clear Neisseria gonorrhoeae in vivo.
Nat Commun, 12, 2021
|
|
1QZC
 
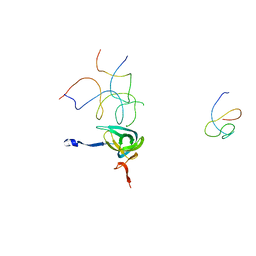 | | Coordinates of S12, SH44, LH69 and SRL separately fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12 | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZB
 
 | | Coordinates of the A-site tRNA model fitted into the cryo-EM map of 70S ribosome in the pre-translocational state | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZD
 
 | | EF-Tu.kirromycin coordinates fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Elongation factor Tu | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZA
 
 | | Coordinates of the A/T site tRNA model fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1R2X
 
 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
4V48
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
5UF6
 
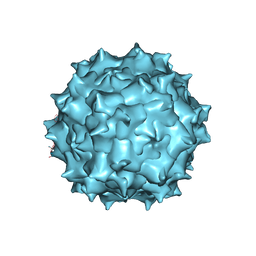 | | The 2.8 A Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, capsid protein VP1 | | Authors: | Xie, Q, Spear, J.M, Noble, A.J, Sousa, D.R, Meyer, N.L, Davulcu, O, Zhang, F, Linhardt, R.J, Stagg, S.M, Chapman, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The 2.8 angstrom Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparinoid Pentasaccharide.
Mol Ther Methods Clin Dev, 5, 2017
|
|
1R2W
 
 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of the 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
8EHG
 
 | | Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
8EMQ
 
 | | Mouse apoferritin heavy chain with zinc determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-28 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (1.66 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
8EN7
 
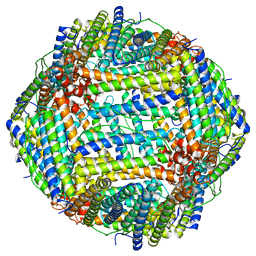 | | Mouse apoferritin heavy chain without zinc determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-28 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (1.68 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
3J4K
 
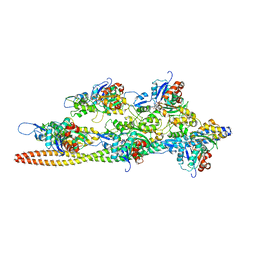 | | Cryo-EM structures of the actin:tropomyosin filament reveal the mechanism for the transition from C- to M-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sousa, D.R, Stagg, S.M, Stroupe, M.E. | | Deposit date: | 2013-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryo-EM Structures of the Actin:Tropomyosin Filament Reveal the Mechanism for the Transition from C- to M-State.
J.Mol.Biol., 425, 2013
|
|
3J1Q
 
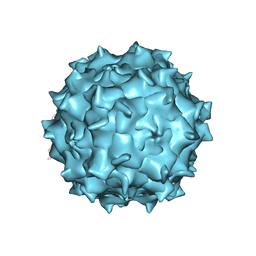 | | Structure of AAV-DJ, a Retargeted Gene Therapy Vector: Cryo-Electron Microscopy at 4.5A resolution | | Descriptor: | Adeno-associated virus DJ | | Authors: | Lerch, T.F, O'Donnell, J.K, Meyer, N.L, Xie, Q, Taylor, K.A, Stagg, S.M, Chapman, M.S. | | Deposit date: | 2012-04-30 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of AAV-DJ, a retargeted gene therapy vector: cryo-electron microscopy at 4.5 A resolution.
Structure, 20, 2012
|
|
8DO6
 
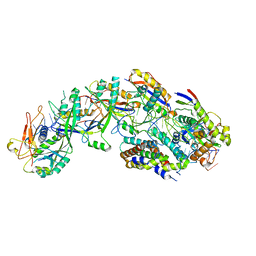 | | The structure of S. epidermidis Cas10-Csm bound to target RNA | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Paraan, M, Stagg, S.M, Dunkle, J.A. | | Deposit date: | 2022-07-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of a Type III-A CRISPR-Cas effector complex reveals conserved and idiosyncratic contacts to target RNA and crRNA among Type III-A systems.
Plos One, 18, 2023
|
|
6WCJ
 
 | | Asymmetric vertex of the clathrin minicoat cage | | Descriptor: | Clathrin heavy chain 1, Clathrin light chain B | | Authors: | Paraan, M, Mendez, J, Sharum, S, Kurtin, D, He, H, Stagg, S. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | The structures of natively assembled clathrin-coated vesicles.
Sci Adv, 6, 2020
|
|
4RDP
 
 | | Crystal structure of Cmr4 | | Descriptor: | CRISPR system Cmr subunit Cmr4 | | Authors: | Shao, Y, Tang, L, Li, H. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Essential Structural and Functional Roles of the Cmr4 Subunit in RNA Cleavage by the Cmr CRISPR-Cas Complex.
Cell Rep, 9, 2014
|
|
6PQB
 
 | |
6NZ0
 
 | | Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2 | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein, MAGNESIUM ION | | Authors: | Meyer, N.L, Xie, Q, Davulcu, O, Yoshioka, C, Chapman, M.S. | | Deposit date: | 2019-02-12 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the gene therapy vector, adeno-associated virus with its cell receptor, AAVR.
Elife, 8, 2019
|
|
6C22
 
 | |
6C21
 
 | |
3J4P
 
 | | Electron Microscopy Analysis of a Disaccharide Analog complex Reveals Receptor Interactions of Adeno-Associated Virus | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Capsid protein VP1, MAGNESIUM ION, ... | | Authors: | Xie, Q, Chapman, M.S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Electron microscopy analysis of a disaccharide analog complex reveals receptor interactions of adeno-associated virus.
J.Struct.Biol., 184, 2013
|
|
