6P04
 
 | |
6P01
 
 | | Apo structure of the E52D mutant of ANT-4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Kanamycin nucleotidyltransferase, ... | | Authors: | Selvaraj, B, Cuneo, M.J. | | Deposit date: | 2019-05-16 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | "Catch and Release": a Variation of the Archetypal Nucleotidyl Transfer Reaction
Acs Catalysis, 10, 2020
|
|
6P08
 
 | |
6P06
 
 | |
6UN8
 
 | | Wild type ANT bound to neomycin | | Descriptor: | Aminoglycoside NucleotidylTransferase, MAGNESIUM ION, NEOMYCIN | | Authors: | Cuneo, M.J, Selvaraj, B. | | Deposit date: | 2019-10-11 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | "Catch and Release": a Variation of the Archetypal NucleotidylTransfer Reaction
ACS Catal, 10, 2020
|
|
8TX6
 
 | |
8TX5
 
 | |
5EQU
 
 | | Crystal structure of the epimerase SnoN in complex with Fe3+, alpha ketoglutarate and nogalamycin RO | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Nogalamycin RO, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-13 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ERL
 
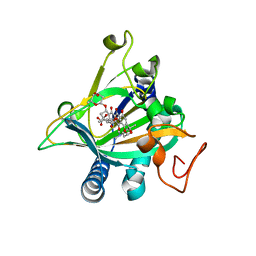 | | Crystal structure of the epimerase SnoN in complex with Ni2+, succinate and nogalamycin RO | | Descriptor: | NICKEL (II) ION, Nogalamycin RO, SUCCINIC ACID, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-14 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EP9
 
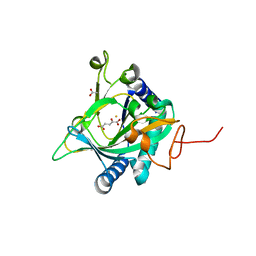 | | Crystal structure of the non-heme alpha ketoglutarate dependent epimerase SnoN from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Niiranen, L, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EPA
 
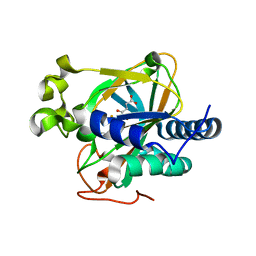 | | Crystal structure of non-heme alpha ketoglutarate dependent carbocyclase SnoK from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Niiranen, L, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MEZ
 
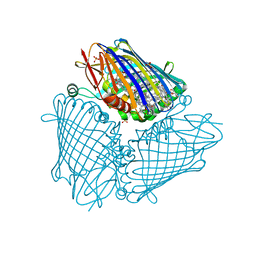 | | X-ray structure of the Fenna-Matthews-Olsen antenna complex from Prosthecochloris aestuarii | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein, SULFATE ION | | Authors: | Selvaraj, B, Lu, X, Cuneo, M.J, Myles, D.A.A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Neutron and X-ray analysis of the Fenna-Matthews-Olson photosynthetic antenna complex from Prosthecochloris aestuarii.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
6NAF
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Selvaraj, B, Park, J, Cuneo, M.J, Myles, D.A.A, Baker, D. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.923 Å), X-RAY DIFFRACTION | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
6NMK
 
 | |
6NMM
 
 | |
6NLT
 
 | | Apo structure of the T130K mutant of ANT-4 | | Descriptor: | CALCIUM ION, Kanamycin nucleotidyltransferase | | Authors: | Selvaraj, B, Cuneo, M.J. | | Deposit date: | 2019-01-09 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catch and release: A novel variation of the archetypal nucleotidyl transfer reaction
To be published
|
|
6NML
 
 | |
6NMN
 
 | |
6MB4
 
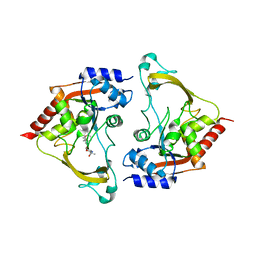 | | Binary (sisomicin) structure of AAC-IIIb | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aac(3)-IIIb protein | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MB6
 
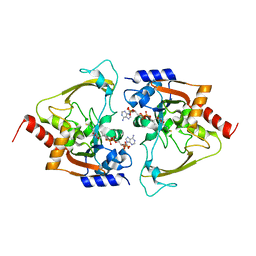 | | AAC-IIIb binary with CoASH | | Descriptor: | Aac(3)-IIIb protein, COENZYME A, MALONATE ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MB5
 
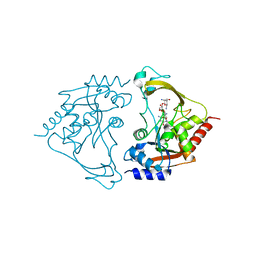 | | AAC-IIIb binary with NEOMYCIN | | Descriptor: | Aac(3)-IIIb protein, NEOMYCIN | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MB7
 
 | | Binary (paromomycin) structure of AAC-IIIb | | Descriptor: | Aac(3)-IIIb protein, PAROMOMYCIN | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MB9
 
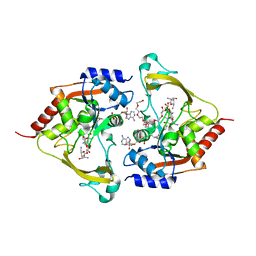 | | Ternary (neomycin/CoA) structure of AAC-IIIb | | Descriptor: | Aac(3)-IIIb protein, COENZYME A, NEOMYCIN, ... | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6MB8
 
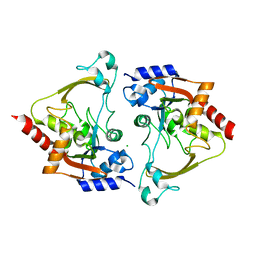 | | Apo structure of AAC-IIIb | | Descriptor: | Aac(3)-IIIb protein, CHLORIDE ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
6N9H
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Park, J, Baker, D. | | Deposit date: | 2018-12-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
