6ZHI
 
 | |
2M7J
 
 | |
2M7I
 
 | |
8OUR
 
 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 16 | | Descriptor: | 5-[1-(3-morpholin-4-yl-1-bicyclo[1.1.1]pentanyl)-2-propan-2-yl-imidazol-4-yl]-3-(trifluoromethyloxy)pyridin-2-amine, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, McEwan, P.A, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
8OUS
 
 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 19 | | Descriptor: | (1S)-1-[4-[6-azanyl-5-(trifluoromethyloxy)pyridin-3-yl]-1-(3-morpholin-4-yl-1-bicyclo[1.1.1]pentanyl)imidazol-2-yl]-2-methyl-propan-1-ol, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, McEwan, P.A, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
8OUT
 
 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 22 | | Descriptor: | (1R)-1-[4-[6-azanyl-5-(trifluoromethyloxy)pyridin-3-yl]-1-(3-fluoranyl-1-bicyclo[1.1.1]pentanyl)imidazol-2-yl]-2,2,2-tris(fluoranyl)ethanol, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, Akkermans, O, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
4PIJ
 
 | | X-ray crystal structure of the K11S/K63S double mutant of ubiquitin | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin | | Authors: | Loll, P.J, Xu, P.J, Schmidt, J, Melideo, S.L. | | Deposit date: | 2014-05-08 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhancing ubiquitin crystallization through surface-entropy reduction.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4PIH
 
 | | X-ray crystal structure of the K33S mutant of ubiquitin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Ubiquitin | | Authors: | Loll, P.J, Xu, P.J, Schmidt, J, Melideo, S.L. | | Deposit date: | 2014-05-08 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhancing ubiquitin crystallization through surface-entropy reduction.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4PIG
 
 | | Crystal structure of the ubiquitin K11S mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Loll, P.J, Xu, P.J, Schmidt, J, Melideo, S.L. | | Deposit date: | 2014-05-08 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Enhancing ubiquitin crystallization through surface-entropy reduction.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
9FKD
 
 | |
3CBG
 
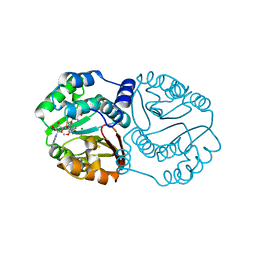 | | Functional and Structural Characterization of a Cationdependent O-Methyltransferase from the Cyanobacterium Synechocystis Sp. Strain PCC 6803 | | Descriptor: | (2E)-3-(3-hydroxy-4-methoxyphenyl)prop-2-enoic acid, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, ... | | Authors: | Kopycki, J.G, Neumann, P, Stubbs, M.T. | | Deposit date: | 2008-02-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Characterization of a Cation-dependent O-Methyltransferase from the Cyanobacterium Synechocystis sp. Strain PCC 6803
J.Biol.Chem., 283, 2008
|
|
7ZAK
 
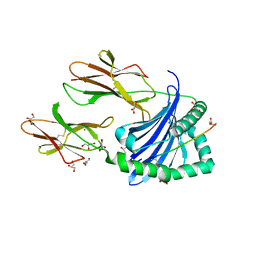 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
8S1X
 
 | |
1PWP
 
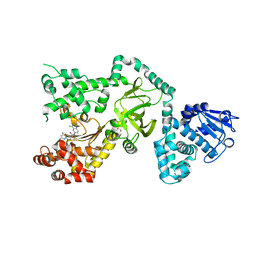 | | Crystal Structure of the Anthrax Lethal Factor complexed with Small Molecule Inhibitor NSC 12155 | | Descriptor: | Lethal factor, N,N'-BIS(4-AMINO-2-METHYLQUINOLIN-6-YL)UREA, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of small molecule inhibitors of anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2O36
 
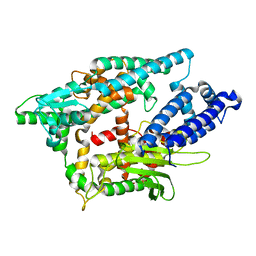 | |
2O3E
 
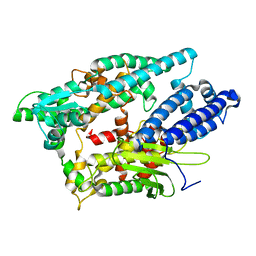 | |
6HPZ
 
 | | Crystal structure of ENL (MLLT1) in complex with acetyllysine | | Descriptor: | 1,2-ETHANEDIOL, N(6)-ACETYLLYSINE, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HPW
 
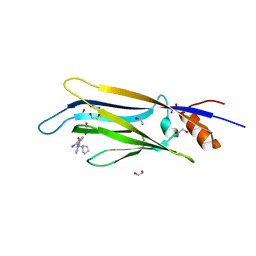 | | Crystal structure of ENL (MLLT1) in complex with compound 20 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HQ0
 
 | | Crystal structure of ENL (MLLT1), apo form | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HPX
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[(3-chlorophenyl)methyl]-1-(2-pyrrolidin-1-ylethyl)benzimidazole-5-carboxamide | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
8T1I
 
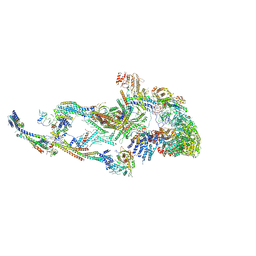 | | Atomic model of the mammalian Mediator complex with MED26 subunit | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
6W1S
 
 | | Atomic model of the mammalian Mediator complex | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Young, N, Asturias, F. | | Deposit date: | 2020-03-04 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers.
Cell, 178, 2019
|
|
8T9D
 
 | | CryoEM structure of TR-TRAP | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
