3RFN
 
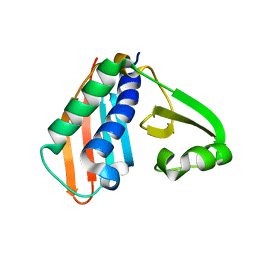 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_1wnu_001, ZINC ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RHU
 
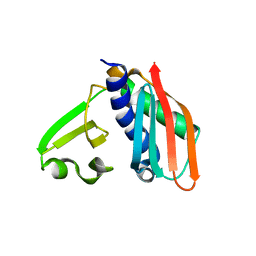 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | SC_1wnu | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RI0
 
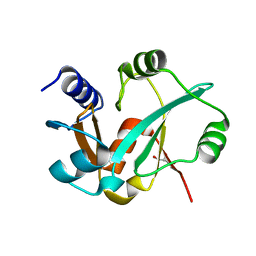 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_2cx5_001, GLYCEROL, SULFATE ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RIJ
 
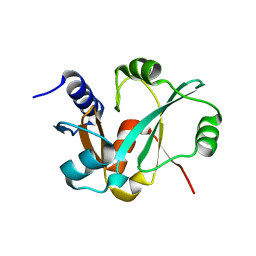 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | GLYCEROL, SC_2cx5 | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
9GBE
 
 | |
6V6G
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V6P
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V8V
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-2 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V83
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-1 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V7T
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase in complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V5E
 
 | | Crystal structure of CTX-M-14 P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6C54
 
 | | Ebola nucleoprotein nucleocapsid-like assembly and the asymmetric unit | | Descriptor: | Nucleoprotein | | Authors: | Su, Z, Wu, C, Pintilie, G.D, Chiu, W, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2018-01-13 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Electron Cryo-microscopy Structure of Ebola Virus Nucleoprotein Reveals a Mechanism for Nucleocapsid-like Assembly.
Cell, 172, 2018
|
|
