6PL0
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris in the Pr state bound to BV chromophore | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Goldbaum, F, Chavas, L, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2019-06-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
3ZG0
 
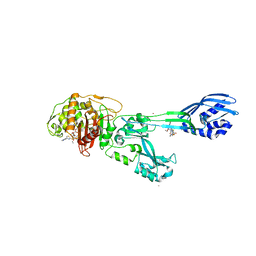 | | Crystal structure of ceftaroline acyl-PBP2a from MRSA with non- covalently bound ceftaroline and muramic acid at allosteric site obtained by cocrystallization | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ceftaroline, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZFZ
 
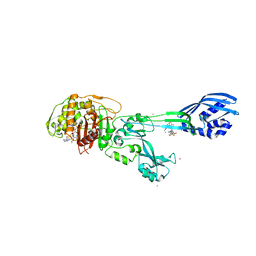 | | Crystal structure of ceftaroline acyl-PBP2a from MRSA with non- covalently bound ceftaroline and muramic acid at allosteric site obtained by soaking | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ceftaroline, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZG5
 
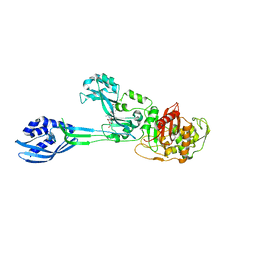 | | Crystal structure of PBP2a from MRSA in complex with peptidoglycan analogue at allosteric | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDOGLYCAN ANALOGUE, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7L5A
 
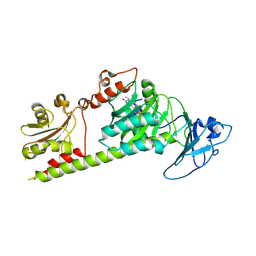 | | Crystal structure of the photosensory module from Xanthomonas campestris bacteriophytochrome XccBphP in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
7L59
 
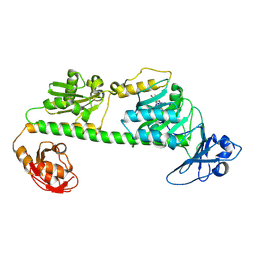 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP-G454E variant from Xanthomonas campestris in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
5AKP
 
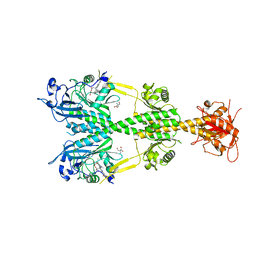 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris bound to BV chromophore | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BILIVERDINE IX ALPHA, CHLORIDE ION, ... | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2015-03-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Full-Length Bacteriophytochrome from the Plant Pathogen Xanthomonas Campestris Provides Clues to its Long-Range Signaling Mechanism.
J.Mol.Biol., 428, 2016
|
|
4OA3
 
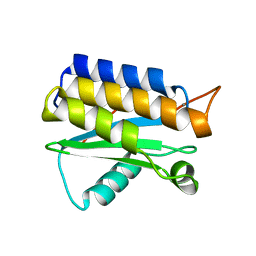 | | Crystal structure of the BA42 protein from BIZIONIA ARGENTINENSIS | | Descriptor: | CALCIUM ION, PROTEIN BA42 | | Authors: | Otero, L.H, Klinke, S, Aran, M, Pellizza, L, Goldbaum, F.A, Cicero, D. | | Deposit date: | 2014-01-03 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain.
Proteins, 82, 2014
|
|
6NDP
 
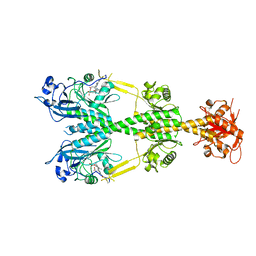 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant L193Q from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Rinaldi, J, Conforte, V, Malamud, F, Goldbaum, F.A, Chavas, L, Bonomi, H.R. | | Deposit date: | 2018-12-14 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
5UYR
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant D199A from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2017-02-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
6NDO
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant L193N from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Rinaldi, J, Conforte, V, Malamud, F, Goldbaum, F.A, Chavas, L, Bonomi, H.R. | | Deposit date: | 2018-12-14 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
5U7N
 
 | | CRYSTAL STRUCTURE OF A CHIMERIC CUA DOMAIN (SUBUNIT II) OF CYTOCHROME BA3 FROM THERMUS THERMOPHILUS WITH THE AMICYANIN LOOP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Cytochrome c oxidase subunit 2 | | Authors: | Otero, L.H, Klinke, S, Espinoza-Cara, A, Vila, A.J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering a bifunctional copper site in the cupredoxin fold by loop-directed mutagenesis.
Chem Sci, 9, 2018
|
|
5ANP
 
 | |
6V95
 
 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-tartaramidoyl derivative (diNGT) | | Descriptor: | (2R,3R)-N-[(1-{(3S,3aR,6S,6aR)-6-[4-({[(2R,3R)-2,3-dihydroxy-4-oxo-4-{[(2R,3R,4R,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]amino}butanoyl]amino}methyl)-1H-1,2,3-triazol-1-yl]hexahydrofuro[3,2-b]furan-3-yl}-1H-1,2,3-triazol-4-yl)methyl]-2,3-dihydroxy-N'-[(2R,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]butanediamide (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-13 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VGF
 
 | | Peanut lectin complexed with divalent S-beta-D-thiogalactopyranosyl beta-D-glucopyranoside derivative (diSTGD) | | Descriptor: | (2S,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6R)-3,4,5-trihydroxy-6-{[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-({4-[({[(2S,3S,4S,5R,6S)-3,4,5-trihydroxy-6-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-2-yl]methyl}sulfanyl)methyl]-1H-1,2,3-triazol-1-yl}methyl)tetrahydro-2H-pyran-2-yl]oxy}tetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VAW
 
 | | Peanut lectin complexed with N-beta-D-galactopyranosyl-L-succinamoyl derivative (NGS) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC4
 
 | | Peanut lectin complexed with S-beta-D-Thiogalactopyranosyl beta-D-glucopyranoside derivative (STGD) | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6S)-3,4,5-trihydroxy-6-methoxytetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VAV
 
 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-succinamoyl derivative (diNGS) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC3
 
 | | Peanut lectin complexed with S-beta-D-thiogalactopyranosyl 6-deoxy-6-S-propynyl-beta-D-glucopyranoside (STG) | | Descriptor: | 6-S-(prop-2-yn-1-yl)-6-thio-beta-D-glucopyranosyl 1-thio-beta-D-galactopyranoside, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4BL2
 
 | |
4BHY
 
 | | Structure of alanine racemase from Aeromonas hydrophila | | Descriptor: | ALANINE RACEMASE | | Authors: | Otero, L.H, Carrasco-Lopez, C, Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2013-04-09 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for the Broad Specificity of a New Family of Amino-Acid Racemases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BL3
 
 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
4CPK
 
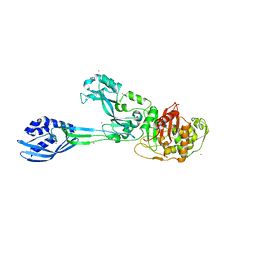 | | Crystal structure of PBP2a double clinical mutant N146K-E150K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, Penicillin binding protein 2 prime, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
4D6Y
 
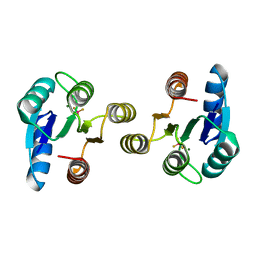 | | Crystal structure of the receiver domain of NtrX from Brucella abortus in complex with beryllofluoride and magnesium | | Descriptor: | BACTERIAL REGULATORY, FIS FAMILY PROTEIN, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Otero, L.H, Fernandez, I, Carrica, M.C, Klinke, S, Goldbaum, F.A. | | Deposit date: | 2014-11-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of Conformational Changes Shed Light Into the Ntrx Receiver Domain Signal Transduction Mechanism
J.Mol.Biol., 427, 2015
|
|
7MHW
 
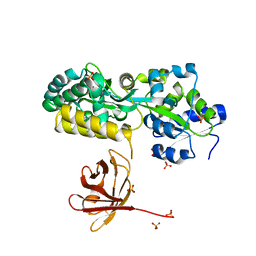 | | Crystal structure of the protease inhibitor U-Omp19 from Brucella abortus fused to Maltose-binding protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Outer membrane lipoprotein omp19, SULFATE ION | | Authors: | Darriba, M.L, Klinke, S, Otero, L.H, Cerutti, M.L, Cassataro, J, Pasquevich, K.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A disordered region retains the full protease inhibitor activity and the capacity to induce CD8 + T cells in vivo of the oral vaccine adjuvant U-Omp19.
Comput Struct Biotechnol J, 20, 2022
|
|
