3J07
 
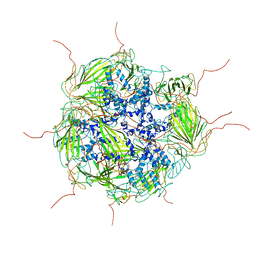 | | Model of a 24mer alphaB-crystallin multimer | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | Deposit date: | 2011-04-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | Cite: | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5OF2
 
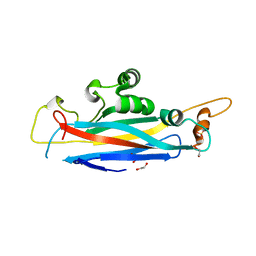 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Spore coat-associated protein N | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1R2N
 
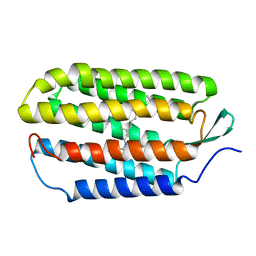 | | NMR structure of the all-trans retinal in dark-adapted Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, terLaak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-09-29 | | Release date: | 2003-10-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6QAY
 
 | |
1A90
 
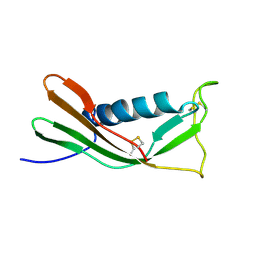 | | RECOMBINANT MUTANT CHICKEN EGG WHITE CYSTATIN, NMR, 31 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-04-14 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
1A67
 
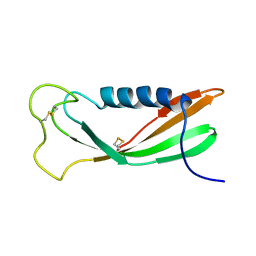 | | CHICKEN EGG WHITE CYSTATIN WILDTYPE, NMR, 16 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-03-06 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
5KGQ
 
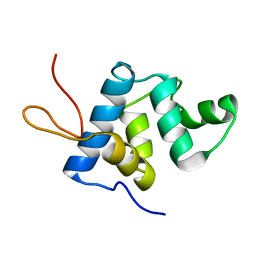 | | NMR structure and dynamics of Q4DY78, a conserved kinetoplasid-specific protein from Trypanosoma cruzi | | Descriptor: | Uncharacterized protein | | Authors: | D'Andrea, E.D, Retel, J.S, Diehl, A, Schmieder, P, Oschkinat, H, Pires, J.R. | | Deposit date: | 2016-06-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of Q4DY78, a conserved kinetoplasid-specific protein from Trypanosoma cruzi.
J.Struct.Biol., 213, 2021
|
|
3O5N
 
 | | Tetrahydroquinoline carboxylates are potent inhibitors of the Shank PDZ domain, a putative target in autism disorders | | Descriptor: | (3aS,4R,9bR)-9-nitro-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-4,6-dicarboxylic acid, SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Saupe, J, Roske, Y, Schillinger, C, Kamdem, N, Radetzki, S, Diehl, A, Oschkinat, H, Krause, G, Heinemann, U, Rademann, J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery, structure-activity relationship studies, and crystal structure of nonpeptide inhibitors bound to the shank3 PDZ domain.
Chemmedchem, 6, 2011
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1MPH
 
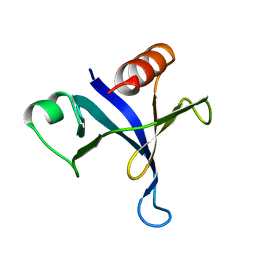 | | PLECKSTRIN HOMOLOGY DOMAIN FROM MOUSE BETA-SPECTRIN, NMR, 50 STRUCTURES | | Descriptor: | BETA SPECTRIN | | Authors: | Nilges, M, Macias, M.J, O'Donoghue, S.I, Oschkinat, H. | | Deposit date: | 1997-04-23 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin.
J.Mol.Biol., 269, 1997
|
|
7ZBQ
 
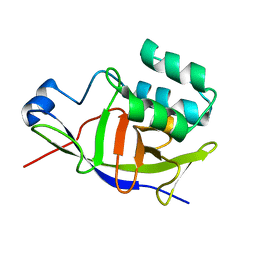 | | Structure of the ADP-ribosyltransferase TccC3HVR from Photorhabdus Luminescens | | Descriptor: | TccC3 | | Authors: | Lindemann, F, Belyy, A, Friedrich, D, Schmieder, P, Bardiaux, B, Roderer, D, Funk, J, Protze, J, Bieling, P, Oschkinat, H, Raunser, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Mechanism of threonine ADP-ribosylation of F-actin by a Tc toxin.
Nat Commun, 13, 2022
|
|
6ZBQ
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 5-fluoranyl-2-(5,6,7,8-tetrahydronaphthalen-2-ylsulfonylamino)benzoic acid, Dishevelled, dsh homolog 3 (Drosophila), ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZBZ
 
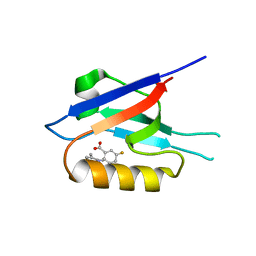 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3,4-dimethylphenyl)sulfonylamino]-5-fluoranyl-benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZC7
 
 | |
6ZC4
 
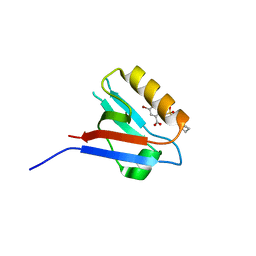 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 5-bromanyl-2-(5,6,7,8-tetrahydronaphthalen-2-ylsulfonylamino)benzoic acid, Dishevelled, dsh homolog 3 (Drosophila), ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZC6
 
 | |
6ZC3
 
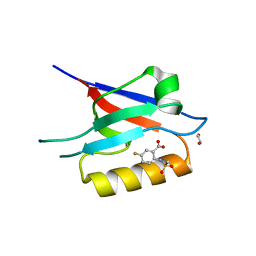 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoranyl-2-(methylsulfonylamino)benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZC8
 
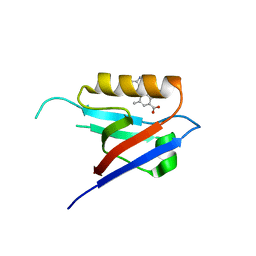 | |
2KLR
 
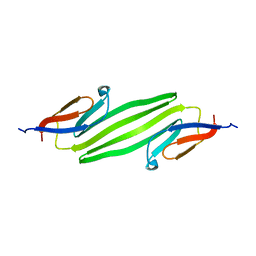 | | Solid-state NMR structure of the alpha-crystallin domain in alphaB-crystallin oligomers | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Rajagopal, P, Markovic, S, Bardiaux, B, Kuehne, R, Higman, V.A, Klevit, R.E, van Rossum, B, Oschkinat, H. | | Deposit date: | 2009-07-08 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1YWJ
 
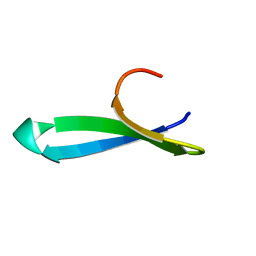 | | Structure of the FBP11WW1 domain | | Descriptor: | Formin-binding protein 3 | | Authors: | Pires, J.R, Parthier, C, Aido-Machado, R, Wiedemann, U, Otte, L, Boehm, G, Rudolph, R, Oschkinat, H. | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain.
J.Mol.Biol., 348, 2005
|
|
1XZ9
 
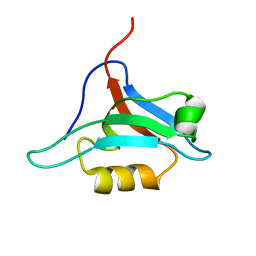 | | Structure of AF-6 PDZ domain | | Descriptor: | Afadin | | Authors: | Joshi, M, Boisguerin, P, Leitner, D, Volkmer-Engert, R, Moelling, K, Schade, M, Schmieder, P, Krause, G, Oschkinat, H. | | Deposit date: | 2004-11-12 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
1YWI
 
 | | Structure of the FBP11WW1 domain complexed to the peptide APPTPPPLPP | | Descriptor: | Formin, Formin-binding protein 3 | | Authors: | Pires, J.R, Parthier, C, Aido-Machado, R, Wiedemann, U, Otte, L, Boehm, G, Rudolph, R, Oschkinat, H. | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain.
J.Mol.Biol., 348, 2005
|
|
1R84
 
 | | NMR structure of the 13-cis-15-syn retinal in dark_adapted bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, Ter Laak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2QG1
 
 | | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1) | | Descriptor: | 1,2-ETHANEDIOL, Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Salah, E, Phillips, C, Savitsky, P, Boisguerin, P, Oschkinat, H, Gileadi, C, Yang, X, Elkins, J.M, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1).
To be Published
|
|
2RQK
 
 | | NMR Solution Structure of Mesoderm Development (MESD) - closed conformation | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Lighthouse, J.K, Werther, T, Andersen, O.M, Diehl, A, Schmieder, P, Holdener, B.C, Oschkinat, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding
Structure, 19, 2011
|
|
