4V4K
 
 | |
8VUZ
 
 | | L5A7 Fab bound to 28H6E11 anti-idiotype Fab | | Descriptor: | 28H6E11 Fab Heavy Chain, 28H6E11 Fab Light Chain, L5A7 Fab Heavy Chain, ... | | Authors: | Olia, A.S, Morano, N.C, Kwong, P.D. | | Deposit date: | 2024-01-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody.
Front Immunol, 15, 2024
|
|
8VUE
 
 | | L5A7 Fab bound to Indonesia2005 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Olia, A.S, Gorman, J, Kwong, P.D. | | Deposit date: | 2024-01-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody.
Front Immunol, 15, 2024
|
|
3LJ5
 
 | |
2POH
 
 | |
8F7T
 
 | | Glycan-Base ConC Env Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env gp120, ... | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-20 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases.
Iscience, 26, 2023
|
|
8VVB
 
 | | Influenza antibody L5A7 Fab | | Descriptor: | L5A7 Heavy Chain, L5A7 Light Chain | | Authors: | Harris, D.R, Olia, A.S, Kwong, P.D. | | Deposit date: | 2024-01-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody.
Front Immunol, 15, 2024
|
|
3C9I
 
 | |
8TDX
 
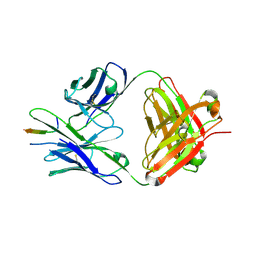 | | TRNM-b.01 in complex with HIV Env fusion peptide | | Descriptor: | Env Fusion Peptide, TRNM-b.01 Fab Heavy Chain, TRNM-b.01 Fab Light Chain | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Broad and potent HIV-1 neutralization in fusion peptide-primed SHIV-boosted macaques
To Be Published
|
|
8F7Z
 
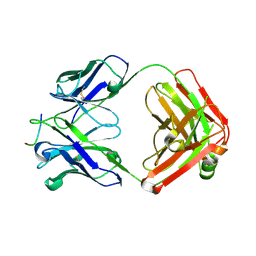 | | VRC34.01_mm28 bound to fusion peptide | | Descriptor: | HIV-1 Env Fusion Peptide, VRC34_m228 Light Chain, VRC34_mm28 Heavy Chain | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
7MTD
 
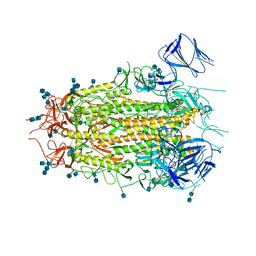 | | Structure of aged SARS-CoV-2 S2P spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tsybovsky, Y, Olia, A.S, Kwong, P.D. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity.
J.Biol.Chem., 297, 2021
|
|
7MTC
 
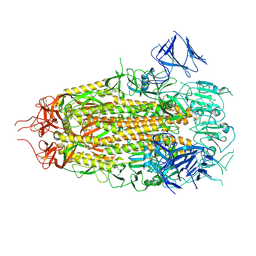 | | Structure of freshly purified SARS-CoV-2 S2P spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tsybovsky, Y, Olia, A.S, Kwong, P.D. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity.
J.Biol.Chem., 297, 2021
|
|
7MTE
 
 | |
8TE7
 
 | | Structure of TRNM-f.01 | | Descriptor: | TRNM-f.01 Fab Heavy Chain, TRNM-f.01 Fab Light Chain | | Authors: | Bender, M.F, Olia, A.S, Kwong, P.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Broad and Potent HIV-1 Neutralization in Fusion Peptide-primed SHIV-boosted Macaques
To Be Published
|
|
5T53
 
 | | MOLECULAR BASIS FOR COHESIN ACETYLATION BY ESTABLISHMENT OF SISTER CHROMATID COHESION N-ACETYLTRANSFERASE ESCO1 | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase ESCO1, ZINC ION | | Authors: | Marmorstein, R, Rivera-Colon, Y, Liszczak, G.P, Olia, A.S, Maguire, A. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Molecular Basis for Cohesin Acetylation by Establishment of Sister Chromatid Cohesion N-Acetyltransferase ESCO1.
J. Biol. Chem., 291, 2016
|
|
6XM0
 
 | | Consensus structure of SARS-CoV-2 spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM5
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-07-29 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM4
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XLU
 
 | | Structure of SARS-CoV-2 spike at pH 4.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
5BU8
 
 | |
7Z7C
 
 | |
8AED
 
 | |
7TXD
 
 | | Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly neutralizing darpin bnd.9, ... | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
