1TYJ
 
 | | Crystal Structure Analysis of type II Cohesin A11 from Bacteroides cellulosolvens | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, cellulosomal scaffoldin | | Authors: | Noach, I, Frolow, F, Jakoby, H, Rosenheck, S, Shimon, L.J.W, Lamed, R, Bayer, E.A. | | Deposit date: | 2004-07-08 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements
J.Mol.Biol., 348, 2005
|
|
3L8Q
 
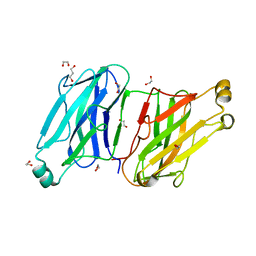 | | Structure analysis of the type II cohesin dyad from the adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2010-01-03 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Modular Arrangement of a Cellulosomal Scaffoldin Subunit Revealed from the Crystal Structure of a Cohesin Dyad
J.Mol.Biol., 399, 2010
|
|
3BWZ
 
 | | Crystal structure of the type II cohesin module from the cellulosome of Acetivibrio cellulolyticus with an extended linker conformation | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Lamed, R, Shimon, L.J.W, Bayer, E, Frolow, F. | | Deposit date: | 2008-01-10 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
1ZV9
 
 | | Crystal structure analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus- SeMet derivative | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETIC ACID, ... | | Authors: | Noach, I, Rosenheck, S, Lamed, R, Shimon, L, Bayer, E, Frolow, F. | | Deposit date: | 2005-06-01 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
3FNK
 
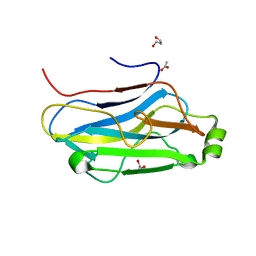 | | Crystal structure of the second type II cohesin module from the cellulosomal adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2008-12-25 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Intermodular Linker Flexibility Revealed from Crystal Structures of Adjacent Cellulosomal Cohesins of Acetivibrio cellulolyticus
J.Mol.Biol., 391, 2009
|
|
3GHP
 
 | |
5FQF
 
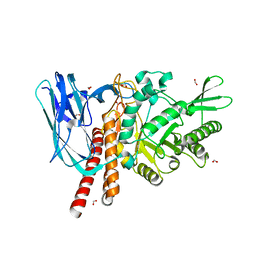 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQH
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQE
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-N-ACETYLGALACTOSAMINIDASE, BROMIDE ION, ... | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FR0
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-deoxy-2-[(difluoroacetyl)amino]-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQG
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5KD2
 
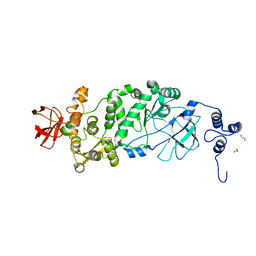 | |
5KDJ
 
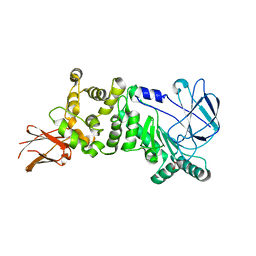 | | ZmpB metallopeptidase from Clostridium perfringens | | Descriptor: | F5/8 type C domain protein, GLYCEROL, SODIUM ION, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDU
 
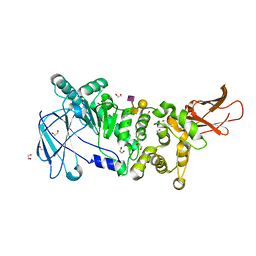 | | ZmpB metallopeptidase in complex with a2,6-Sialyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, SERINE, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDN
 
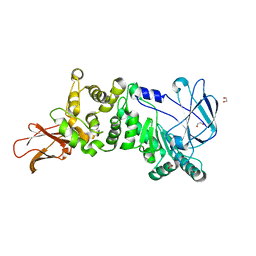 | | ZmpB metallopeptidase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, ZINC ION | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KD8
 
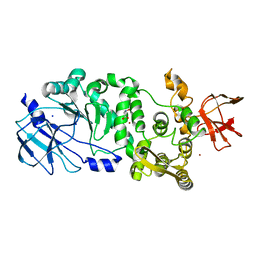 | | BT_4244 metallopeptidase in complex with Tn antigen. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Metallopeptidase, NICKEL (II) ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-07 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDV
 
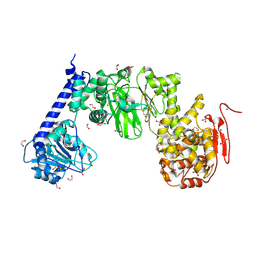 | | IMPa metallopeptidase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, SULFATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDX
 
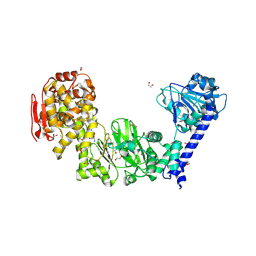 | | IMPa metallopeptidase in complex with T-antigen | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, PHOSPHATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDW
 
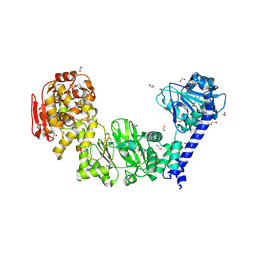 | | IMPa metallopeptidase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, PHOSPHATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KD5
 
 | |
5KDS
 
 | | ZmpB metallopeptidase in complex with an O-glycopeptide (a2,6-sialylated core-3 pentapeptide). | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7JTV
 
 | |
1QZN
 
 | | Crystal Structure Analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus | | Descriptor: | cellulosomal scaffoldin adaptor protein B | | Authors: | Frolow, F, Noach, I, Rosenheck, S, Lamed, R, Qi, X, Shimon, L.J.W, Bayer, E.A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements.
J.Mol.Biol., 348, 2005
|
|
2XDH
 
 | | Non-cellulosomal cohesin from the hyperthermophilic archaeon Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, COHESIN, MAGNESIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Noach, I, Borovok, I, Kwiat, M, Rosenheck, S, Shimon, L.J.W, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Non-Cellulosomal Cohesin from the Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Proteins, 79, 2011
|
|
2ZF9
 
 | |
