2D32
 
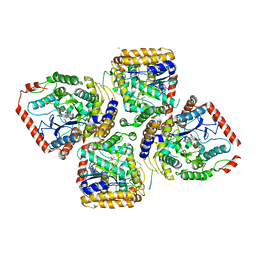 | | Crystal Structure of Michaelis Complex of gamma-Glutamylcysteine Synthetase | | Descriptor: | CYSTEINE, GLUTAMIC ACID, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nakayama, M, Nii, H, Kurokawa, Y, Katano, H, Oda, J. | | Deposit date: | 2005-09-25 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of efficient coupling peptide ligation and ATP hydrolysis by gamma-gluatamylcysteine synthetase
To be Published
|
|
2D33
 
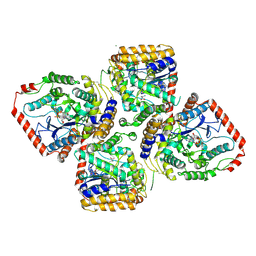 | | Crystal Structure of gamma-Glutamylcysteine Synthetase Complexed with Aluminum Fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CYSTEINE, ... | | Authors: | Hibi, T, Nakayama, M, Nii, H, Kurokawa, Y, Katano, H, Oda, J. | | Deposit date: | 2005-09-25 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of efficient coupling between peptide ligation and ATP hydrolysis by gamma-gluatamylcysteine synthetase
To be Published
|
|
1V4G
 
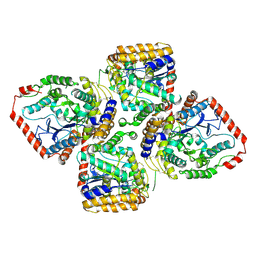 | | Crystal Structure of gamma-Glutamylcysteine Synthetase from Escherichia coli B | | Descriptor: | Glutamate--cysteine ligase | | Authors: | Hibi, T, Nii, H, Nakatsu, T, Kato, H, Hiratake, J, Oda, J. | | Deposit date: | 2003-11-13 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
1VA6
 
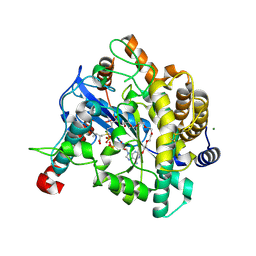 | | Crystal structure of Gamma-glutamylcysteine synthetase from Escherichia Coli B complexed with Transition-state analogue | | Descriptor: | (2S)-2-AMINO-4-[[(2R)-2-CARBOXYBUTYL](PHOSPHONO)SULFONIMIDOYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nii, H, Nakatsu, T, Kato, H, Hiratake, J, Oda, J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
7X7O
 
 | | SARS-CoV-2 spike RBD in complex with neutralizing antibody UT28K | | Descriptor: | Spike protein S1, UT28K Fab, heavy chain, ... | | Authors: | Ozawa, T, Tani, H, Anraku, Y, Kita, S, Igarashi, E, Saga, Y, Inasaki, N, Kawasuji, H, Yamada, H, Sasaki, S, Somekawa, M, Sasaki, J, Hayakawa, Y, Yamamoto, Y, Morinaga, Y, Kurosawa, N, Isobe, M, Fukuhara, H, Maenaka, K, Hashiguchi, T, Kishi, H, Kitajima, I, Saito, S, Niimi, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Novel super-neutralizing antibody UT28K is capable of protecting against infection from a wide variety of SARS-CoV-2 variants.
Mabs, 14, 2022
|
|
3ALE
 
 | |
8K5H
 
 | | Structure of the SARS-CoV-2 BA.1 spike with UT28-RD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Rational in silico design identifies two mutations that restore UT28K SARS-CoV-2 monoclonal antibody activity against Omicron BA.1.
Structure, 32, 2024
|
|
8K5G
 
 | | Structure of the SARS-CoV-2 BA.1 RBD with UT28-RD | | Descriptor: | Spike protein S1, UT28K-RD Fab Heavy chain, UT28K-RD Fab Light chain | | Authors: | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Rational in silico design identifies two mutations that restore UT28K SARS-CoV-2 monoclonal antibody activity against Omicron BA.1.
Structure, 32, 2024
|
|
