1PRA
 
 | |
7AH1
 
 | | L19 diabody fragment from immunocytokine L19-IL2 | | Descriptor: | Anti-(ED-B) scFV | | Authors: | Ongaro, T, Guarino, S.R, Scietti, L, Palamini, M, Wulhfard, S, Villa, A, Neri, D, Forneris, F. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inference of molecular structure for characterization and improvement of clinical grade immunocytokines.
J.Struct.Biol., 213, 2021
|
|
8AS0
 
 | | PD-1 extracellular domain in complex with Fab fragment from D12 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ongaro, T, Scietti, L, Pluss, L, Peissert, F, Villa, A, Puca, E, De Luca, R, Neri, D, Forneris, F. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Selection of a PD-1 blocking antibody from a novel fully human phage display library.
Protein Sci., 31, 2022
|
|
8ILV
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 18 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[3-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILS
 
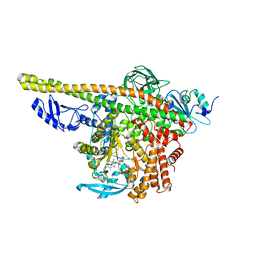 | | Cryo-EM structure of PI3Kalpha in complex with compound 17 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2FNB
 
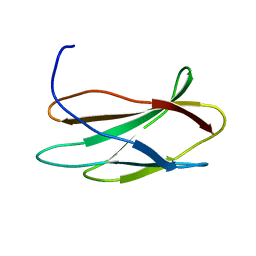 | | NMR STRUCTURE OF THE FIBRONECTIN ED-B DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Fattorusso, R, Pellecchia, M, Viti, F, Neri, P, Neri, D, Wuthrich, K. | | Deposit date: | 1998-12-16 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human oncofoetal fibronectin ED-B domain, a specific marker for angiogenesis.
Structure Fold.Des., 7, 1999
|
|
2X7O
 
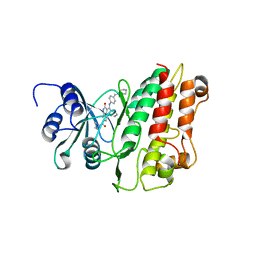 | | Crystal structure of TGFbRI complexed with an indolinone inhibitor | | Descriptor: | (3Z)-N-ETHYL-N-METHYL-2-OXO-3-(PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]AMINO}METHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-6-CARBOXAMIDE, TGF-BETA RECEPTOR TYPE I | | Authors: | Roth, G.J, Heckel, A, Brandl, T, Grauert, M, Hoerer, S, Kley, J.T, Schnapp, G, Baum, P, Mennerich, D, Schnapp, A, Park, J.E. | | Deposit date: | 2010-03-03 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design, Synthesis and Evaluation of Indolinones as Inhibitors of the Transforming Growth Factor Beta Receptor I (Tgfbri)
J.Med.Chem., 53, 2010
|
|
8AYG
 
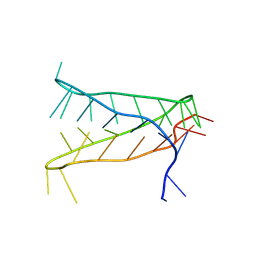 | | Crystal structure of an intramolecular i-motif at the insulin-linked polymorphic region (ILPR) | | Descriptor: | Insulin-linked polymorphic region, ILPR DNA (31-MER) | | Authors: | Parkinson, G.N, Alexandrou, E, Waller, Z.A.E, El-Omari, K. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into i-motif DNA structures in sequences from the insulin-linked polymorphic region.
Nat Commun, 15, 2024
|
|
2R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
