5T62
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3-Tif6-Lsg1 Complex | | Descriptor: | 5.8S Ribosomal RNA, 5S Ribosomal RNA, 60S ribosomal export protein NMD3, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
5T6R
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
7K8T
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C002 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C002 Fab Heavy Chain, ... | | Authors: | Malyutin, A.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
8GHZ
 
 | | Cryo-EM structure of fish immunogloblin M-Fc | | Descriptor: | Teleost immunoglobulin M protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lyu, M, Stadtmueller, B.M, Malyutin, A.G. | | Deposit date: | 2023-03-13 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | The structure of the teleost Immunoglobulin M core provides insights on polymeric antibody evolution, assembly, and function.
Nat Commun, 14, 2023
|
|
7JG1
 
 | | Dimeric Immunoglobin A (dIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
7JG2
 
 | | Secretory Immunoglobin A (SIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
7K8S
 
 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C002 (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C002 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Malyutin, A.G, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8U
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C104 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C104 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Malyutin, A.G, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7T3H
 
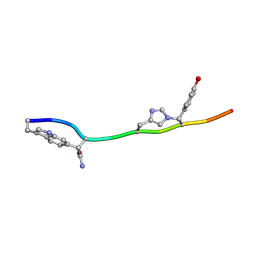 | | MicroED structure of Dynobactin | | Descriptor: | TRP-ASN-SER-ASN-VAL-HIS-SER-TYR-ARG-PHE | | Authors: | Yoo, B.-K, Kaiser, J.T, Rees, D.C, Miller, R.D, Iinishi, A, Lewis, K, Bowman, S. | | Deposit date: | 2021-12-07 | | Release date: | 2022-10-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
7R1W
 
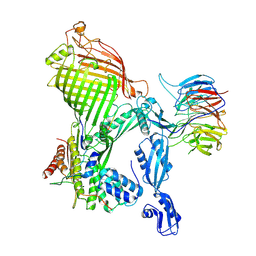 | | E. coli BAM complex (BamABCDE) bound to dynobactin A | | Descriptor: | Dynobactin A, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Hiller, S, Maier, T. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
7R1V
 
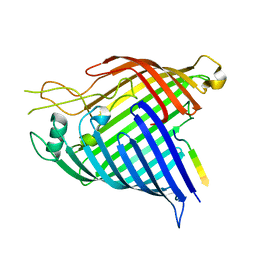 | |
7K8N
 
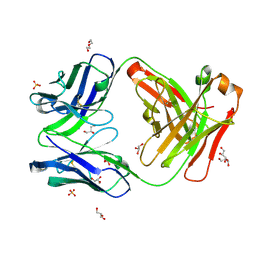 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C102 | | Descriptor: | C102 Fab Heavy Chain, C102 Fab Light Chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8X
 
 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C121 Fab Heavy chain, C121 Fab Light chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Q
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C121 | | Descriptor: | C121 Fab Heavy Chain, C121 Fab Light Chain, GLYCEROL | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Y
 
 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C121 Fab Heavy chain, C121 Fab Light chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8P
 
 | |
7K8M
 
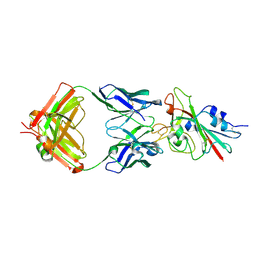 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C102 Fab Heavy Chain, C102 Fab Light Chain, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8V
 
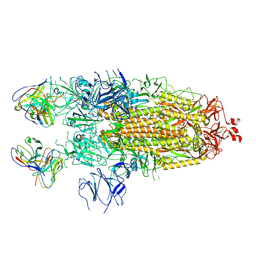 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C110 Fab Heavy Chain, ... | | Authors: | Dam, K.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8O
 
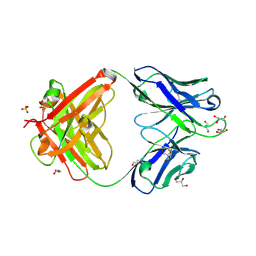 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C002 | | Descriptor: | C002 Fab Heavy Chain, C002 Fab Light Chain, GLYCEROL, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8W
 
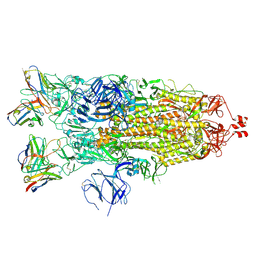 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C119 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C119 Fab Heavy Chain, ... | | Authors: | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8R
 
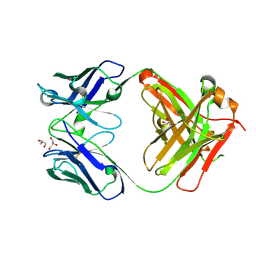 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C135 | | Descriptor: | C135 Fab Heavy Chain, C135 Fab Light Chain, GLYCEROL | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Z
 
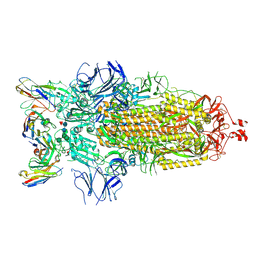 | |
7K90
 
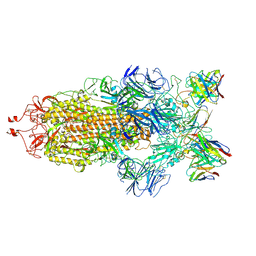 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C144 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C144 Fab Heavy Chain, C144 Fab Light Chain, ... | | Authors: | Barnes, C.O, Esswein, S.R, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7L5W
 
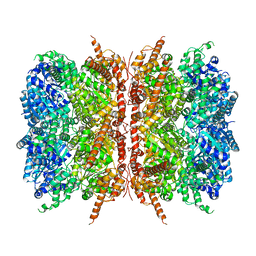 | | p97-R155H mutant dodecamer I | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7L5X
 
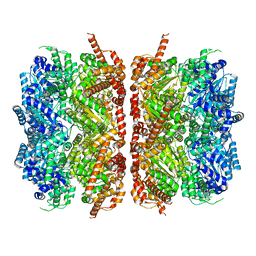 | | p97-R155H mutant dodecamer II | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
