4WCT
 
 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2014-09-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
4OY9
 
 | | Crystal structure of human P-Cadherin EC1-EC2 in closed conformation | | Descriptor: | CALCIUM ION, Cadherin-3 | | Authors: | Dalle Vedove, A, Lucarelli, A.P, Nardone, V, Matino, A, Parisini, E. | | Deposit date: | 2014-02-11 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The X-ray structure of human P-cadherin EC1-EC2 in a closed conformation provides insight into the type I cadherin dimerization pathway.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XWZ
 
 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus in complex with the substrate fructosyl lysine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase, LYSINE, ... | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2015-01-29 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
4ZT1
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in x-dimer conformation | | Descriptor: | CALCIUM ION, Cadherin-1 | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
4ZTE
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in complex with a peptidomimetic inhibitor | | Descriptor: | CALCIUM ION, Cadherin-1, N-{[(2S,5S)-1-benzyl-5-(2-{[(2S,3S)-1-(tert-butylamino)-3-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl)-3,6-dioxopiperazin-2-yl]methyl}-L-alpha-asparagine | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
4AUT
 
 | | Crystal structure of the tuberculosis drug target Decaprenyl- Phosphoryl-beta-D-Ribofuranose-2-oxidoreductase (DprE1) from Mycobacterium smegmatis | | Descriptor: | DECAPRENYL-PHOSPHORYL-BETA-D-RIBOFURANOSE-2-OXIDOREDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Neres, J, Pojer, F, Molteni, E, Chiarelli, L.R, Dhar, N, Boy-Rottger, S, Buroni, S, Fullam, E, Degiacomi, G, Lucarelli, A, Read, R.J, Zanoni, G, Edmondson, D.E, De Rossi, E, Pasca, M, Riccardi, G, Mattevi, A, Dyson, P.J, Cole, S.T, Binda, C. | | Deposit date: | 2012-05-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Benzothiazinone-Mediated Killing of Mycobacterium Tuberculosis.
Sci. Transl. Med., 4, 2012
|
|
4F4Q
 
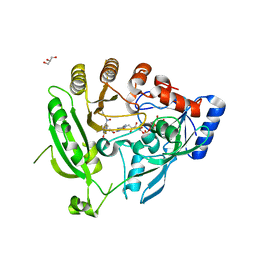 | | Crystal structure of M. smegmatis DprE1 in complex with FAD and covalently bound BTZ043 | | Descriptor: | 8-(hydroxyamino)-2-[(2S)-2-methyl-1,4-dioxa-8-azaspiro[4.5]dec-8-yl]-6-(trifluoromethyl)-4H-1,3-benzothiazin-4-one, DprE1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Neres, J, Pojer, F, Molteni, E, Chiarelli, L, Riccardi, G, Mattevi, A, Cole, S.T, Binda, C. | | Deposit date: | 2012-05-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Structural Basis for Benzothiazinone-Mediated Killing of Mycobacterium tuberculosis.
Sci Transl Med, 4, 2012
|
|
2WZW
 
 | | Crystal structure of the FMN-dependent nitroreductase NfnB from Mycobacterium smegmatis in complex with NADPH | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NFNB PROTEIN, ... | | Authors: | Bellinzoni, M, Manina, G, Riccardi, G, Alzari, P.M. | | Deposit date: | 2009-12-03 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Characterization of the Mycobacterium Smegmatis Nitroreductase Nfnb, and its Role in Benzothiazinone Resistance
Mol.Microbiol., 77, 2010
|
|
2WZV
 
 | | Crystal structure of the FMN-dependent nitroreductase NfnB from Mycobacterium smegmatis | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, NFNB PROTEIN, ... | | Authors: | Bellinzoni, M, Manina, G, Riccardi, G, Alzari, P.M. | | Deposit date: | 2009-12-03 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biological and Structural Characterization of the Mycobacterium Smegmatis Nitroreductase Nfnb, and its Role in Benzothiazinone Resistance
Mol.Microbiol., 77, 2010
|
|
6F8W
 
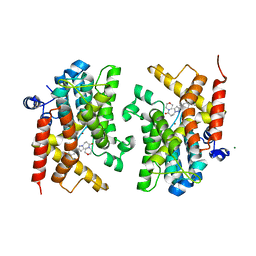 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-18a | | Descriptor: | 3-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F6U
 
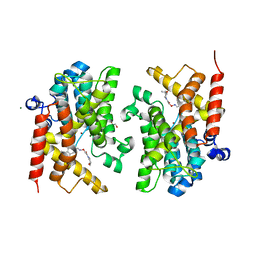 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-7b | | Descriptor: | 2-[(~{E})-(3-cyclopentyloxy-4-methoxy-phenyl)methylideneamino]oxy-1-[(2~{R},6~{S})-2,6-dimethylmorpholin-4-yl]ethanone, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F8T
 
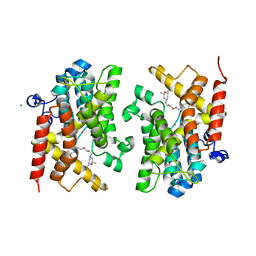 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-4a | | Descriptor: | (2~{R})-1-[(~{E})-(3-cyclopentyloxy-4-methoxy-phenyl)methylideneamino]oxy-3-[(2~{R},6~{S})-2,6-dimethylmorpholin-4-yl]propan-2-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F8V
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-18b | | Descriptor: | 3-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-1-[(2~{R},6~{R})-2,6-dimethylmorpholin-4-yl]propan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F8U
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-20b | | Descriptor: | 2-[(~{E})-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylideneamino]oxy-1-[(2~{R},6~{R})-2,6-dimethylmorpholin-4-yl]ethanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F8X
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-26g | | Descriptor: | 1,2-ETHANEDIOL, 2-[(5~{R})-3-(3-cyclopentyloxy-4-methoxy-phenyl)-4,5-dihydro-1,2-oxazol-5-yl]-~{N},~{N}-bis(2-hydroxyethyl)ethanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F8R
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-54 | | Descriptor: | (2~{S})-1-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6FDC
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-32a | | Descriptor: | (2~{R})-1-[3-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, (2~{S})-1-[5-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
