4V14
 
 | |
8XKV
 
 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Arabidopsis in Apo state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ATP-dependent zinc metalloprotease FTSH 12, chloroplastic, ... | | Authors: | Liang, K, Zhan, X, Xu, Q, Wu, J, Yan, Z. | | Deposit date: | 2023-12-25 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the chloroplast protein import in land plants.
Cell, 187, 2024
|
|
8XQW
 
 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Chlamydomonas reinhardtii in AMPPNP bound state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ACP, ... | | Authors: | Liang, K, Zhan, X, Wu, J, Yan, Z. | | Deposit date: | 2024-01-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conservation and specialization of the Ycf2-FtsHi chloroplast protein import motor in green algae.
Cell, 2024
|
|
8XKU
 
 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Arabidopsis in ATP-bound state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent zinc metalloprotease FTSH 12, ... | | Authors: | Liang, K, Zhan, X, Xu, Q, Wu, J, Yan, Z. | | Deposit date: | 2023-12-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the chloroplast protein import in land plants.
Cell, 187, 2024
|
|
8XQX
 
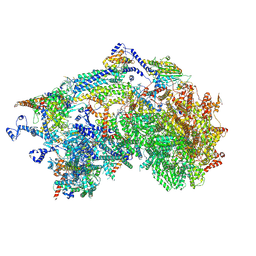 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from chlamydomonas reinhardtii in apo state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ACP, ... | | Authors: | Liang, K, Zhan, X, Wu, J, Yan, Z. | | Deposit date: | 2024-01-05 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conservation and specialization of the Ycf2-FtsHi chloroplast protein import motor in green algae.
Cell, 2024
|
|
5ZQZ
 
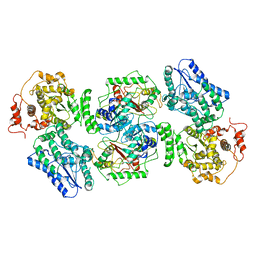 | | Structure of human mitochondrial trifunctional protein, tetramer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZRV
 
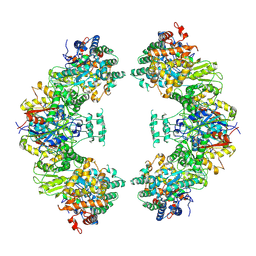 | | Structure of human mitochondrial trifunctional protein, octamer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4TRT
 
 | |
7YOC
 
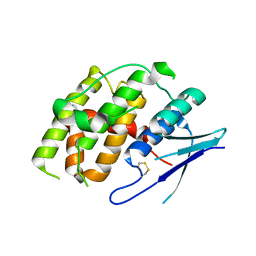 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7CGE
 
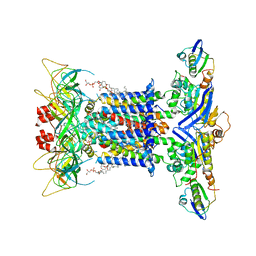 | | The overall structure of nucleotide free MlaFEDB complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7CGN
 
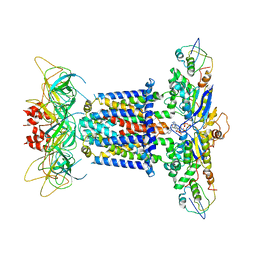 | | The overall structure of the MlaFEDB complex in ATP-bound EQtall conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7CH0
 
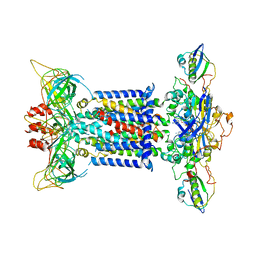 | | The overall structure of the MlaFEDB complex in ATP-bound EQclose conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7OBN
 
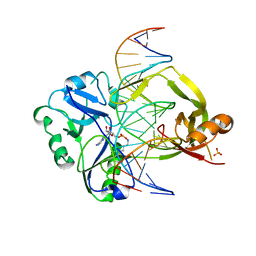 | | Structural investigations of a new L3 DNA ligase: structure-function analysis | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), DNA ligase, ... | | Authors: | Leiros, H.-K.S, Williamson, A. | | Deposit date: | 2021-04-23 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Bacteriophage origin of some minimal ATP-dependent DNA ligases: a new structure from Burkholderia pseudomallei with striking similarity to Chlorella virus ligase.
Sci Rep, 11, 2021
|
|
7YP0
 
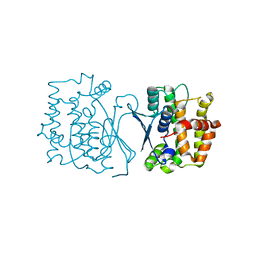 | | Crystal structure of CtGST | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2022-08-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8K2P
 
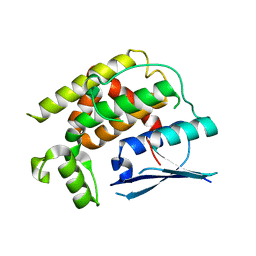 | | Crystal structure of CtGST-F76A | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8K2O
 
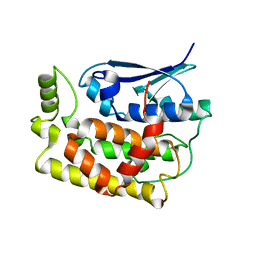 | | Crystal structure of Fhb7-M10 | | Descriptor: | Fhb7-M10 | | Authors: | Yang, J, Lei, X.G, Xiao, J.Y. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8XB3
 
 | |
8XB4
 
 | |
8XB2
 
 | | Structure of radafaxine-bound state of the human Norepinephrine Transporter | | Descriptor: | (2~{S},3~{S})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholin-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GFP-MBP-solute carrier family 6 member 2,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter,Maltose/maltodextrin-binding periplasmic protein,Sodium-dependent noradrenaline transporter | | Authors: | Wu, J.X, Ji, W.M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Substrate binding and inhibition mechanism of norepinephrine transporter.
Nature, 633, 2024
|
|
5JDA
 
 | | Bacillus cereus CotH kinase plus Mg2+/AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Sreelatha, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Phosphorylation of spore coat proteins by a family of atypical protein kinases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JD9
 
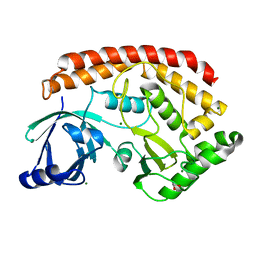 | | Bacillus cereus CotH kinase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Spore coat protein H | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Sreelatha, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Phosphorylation of spore coat proteins by a family of atypical protein kinases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1ODH
 
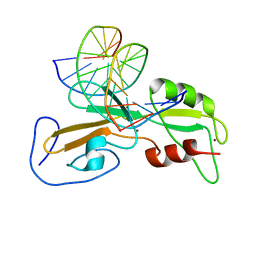 | | Structure of the GCM domain bound to DNA | | Descriptor: | 5'-D(*CP*GP*AP*TP*GP*CP*GP*GP*GP*TP *GP*CP*A)-3', 5'-D(*TP*GP*CP*AP*CP*CP*CP*GP*CP*AP *TP*CP*G)-3', MGCM1, ... | | Authors: | Cohen, S.X, Muller, C.W. | | Deposit date: | 2003-02-19 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of the Gcm Domain-DNA Complex: A DNA-Binding Domain with a Novel Fold and Mode of Target Site Recognition
Embo J., 22, 2003
|
|
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | Descriptor: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | Authors: | Shi, Y, Huang, G, Zhan, X. | | Deposit date: | 2021-07-31 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
7CUN
 
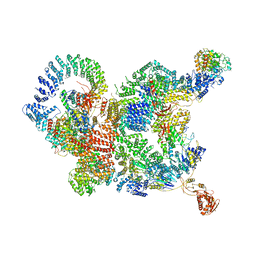 | | The structure of human Integrator-PP2A complex | | Descriptor: | Integrator complex subunit 1, Integrator complex subunit 11, Integrator complex subunit 2, ... | | Authors: | Zheng, H, Qi, Y, Liu, W, Li, J, Wang, J, Xu, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Science, 370, 2020
|
|
7FIL
 
 | |
