5GRM
 
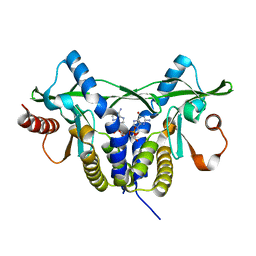 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
5GS5
 
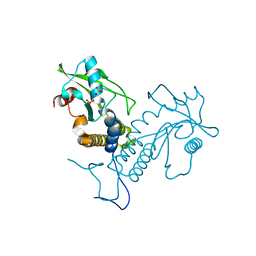 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
3V6L
 
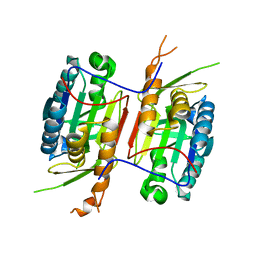 | | Crystal Structure of caspase-6 inactivation mutation | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
3V6M
 
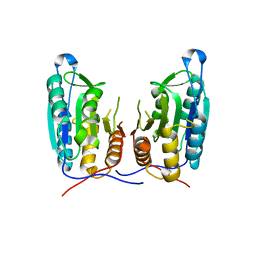 | | Inhibition of caspase-6 activity by single mutation outside the active site | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
2RI0
 
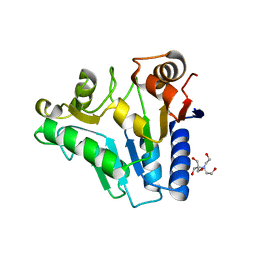 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucosamine-6-phosphate deaminase, SODIUM ION | | Authors: | Li, D, Liu, C, Li, L.F, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
4E2A
 
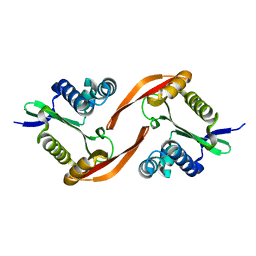 | |
3EXT
 
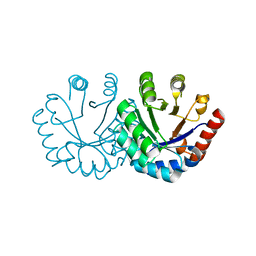 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | MAGNESIUM ION, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Liu, X, Li, G.L, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXS
 
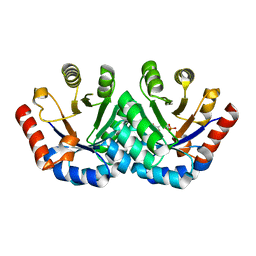 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3H6X
 
 | |
3DEZ
 
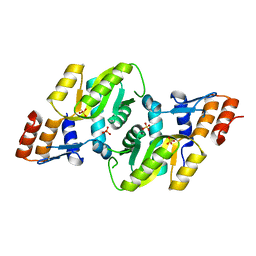 | | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus mutans | | Descriptor: | Orotate phosphoribosyltransferase, SULFATE ION | | Authors: | Liu, C.P, Gao, Z.Q, Hou, H.F, Li, L.F, Su, X.D, Dong, Y.H. | | Deposit date: | 2008-06-11 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of orotate phosphoribosyltransferase from the caries pathogen Streptococcus mutans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2HCU
 
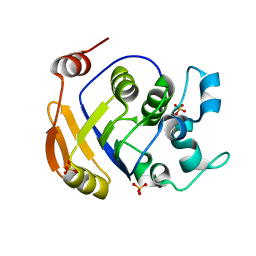 | | Crystal Structure Of Smu.1381 (or LeuD) from Streptococcus Mutans | | Descriptor: | 3-isopropylmalate dehydratase small subunit, SULFATE ION | | Authors: | Gao, Z.Q, Hou, H.F, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-06-19 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure Of Smu.1381 (or LeuD) from Streptococcus Mutans
To be Published
|
|
3B3D
 
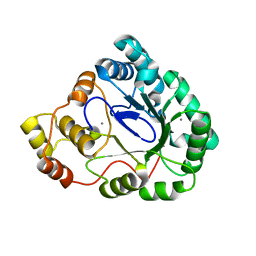 | | B.subtilis YtbE | | Descriptor: | CALCIUM ION, Putative morphine dehydrogenase | | Authors: | Zhou, Y.F, Li, L.F, Liang, Y.H, Su, X.-D. | | Deposit date: | 2007-10-20 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
2HVV
 
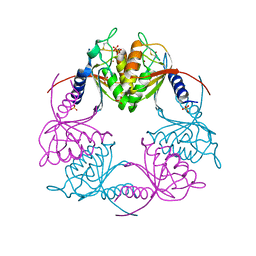 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | Descriptor: | SULFATE ION, ZINC ION, deoxycytidylate deaminase | | Authors: | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
2HVW
 
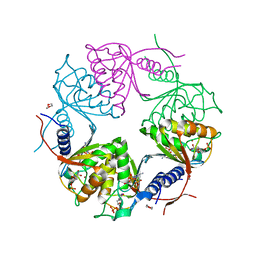 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 3,4-DIHYDRO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
4EGS
 
 | |
3MXZ
 
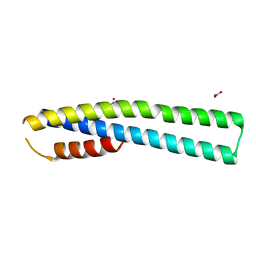 | | Crystal Structure of tubulin folding cofactor A from Arabidopsis thaliana | | Descriptor: | NITRATE ION, Tubulin-specific chaperone A | | Authors: | Lu, L, Nan, J, Mi, W, Su, X.D, Li, Y. | | Deposit date: | 2010-05-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Crystal structure of tubulin folding cofactor A from Arabidopsis thaliana and its beta-tubulin binding characterization
Febs Lett., 584, 2010
|
|
2D4G
 
 | |
3D3F
 
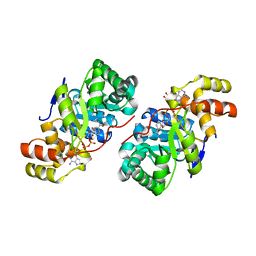 | |
8ITE
 
 | |
3URG
 
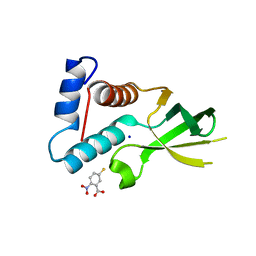 | | The crystal structure of Anabaena CcbP | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Alr1010 protein, SODIUM ION | | Authors: | Fan, X.X, Liu, X, Su, X.D. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Ellman's reagent in promoting crystallization and structure determination of Anabaena CcbP.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2RI1
 
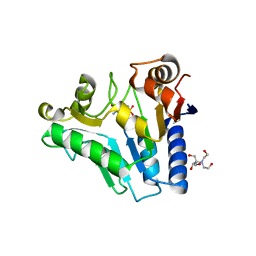 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) with GlcN6P from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine-6-phosphate deaminase | | Authors: | Liu, C, Li, D, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
8YGG
 
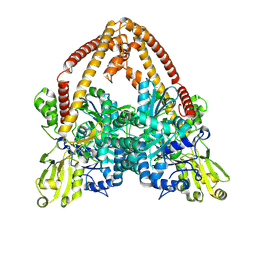 | | pP1192R-apo Closed state | | Descriptor: | DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YGH
 
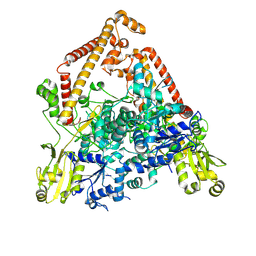 | | pP1192R-apo open state | | Descriptor: | DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YIK
 
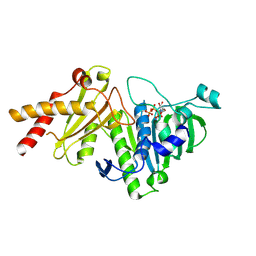 | | pP1192R-ATPase-domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
