4PDW
 
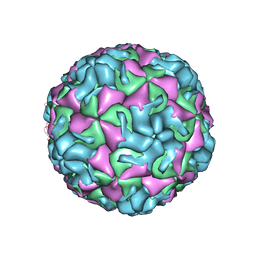 | | A benzonitrile analogue inhibits rhinovirus replication | | Descriptor: | 4-[(4,5-dimethoxy-2-nitrophenyl)acetyl]benzonitrile, Capsid protein VP4/VP2, GLYCEROL, ... | | Authors: | Querol-Audi, J, Guerra, P, Lacroix, C, Roche, M, Franco, D, Froeyen, M, Terme, T, Vanelle, P, Neyts, J, Leyssen, P, Verdaguer, N. | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel benzonitrile analogue inhibits rhinovirus replication.
J.Antimicrob.Chemother., 69, 2014
|
|
6DIZ
 
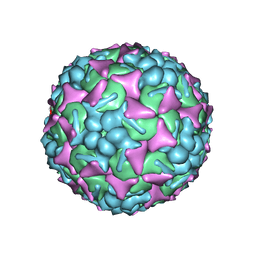 | | EV-A71 strain 11316 complexed with tryptophan dendrimer MADAL_0385 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Sun, L, Lee, H, Thibaut, H.J, Rivero-Buceta, E, Martinez-Gualda, B, Delang, L, Leyssen, P, Gago, F, San-Felix, A, Hafenstein, S, Mirabelli, C, Neyts, J. | | Deposit date: | 2018-05-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Viral engagement with host (co-)receptors blocked by a novel class of tryptophan dendrimers that targets the 5-fold-axis of the enterovirus-A71 capsid.
Plos Pathog., 15, 2019
|
|
5ABJ
 
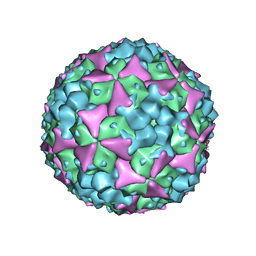 | | Structure of Coxsackievirus A16 in complex with GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
8PQ2
 
 | | XBB 1.0 RBD bound to P4J15 (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, P4J15 Fragment Antigen-Binding Heavy Chain, P4J15 Fragment Antigen-Binding Light Chain, ... | | Authors: | Duhoo, Y, Lau, K. | | Deposit date: | 2023-07-10 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
8PSD
 
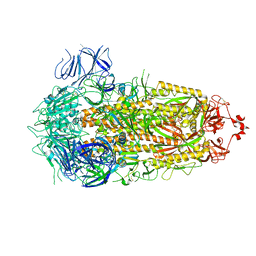 | | SARS-CoV-2 XBB 1.0 closed conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Duhoo, Y, Lau, K. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
7AEG
 
 | | SARS-CoV-2 main protease in a covalent complex with SDZ 224015 derivative, compound 5 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-[(1S)-1-(carboxymethyl)-3-fluoro-2-oxopropyl]-L-alaninamide | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
7AEH
 
 | | SARS-CoV-2 main protease in a covalent complex with a pyridine derivative of ABT-957, compound 1 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
7B3E
 
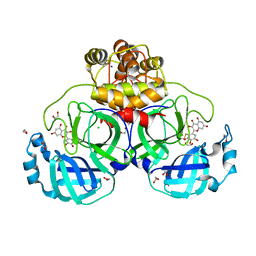 | | Crystal structure of myricetin covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-11-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Acs Pharmacol Transl Sci, 4, 2021
|
|
3ZVG
 
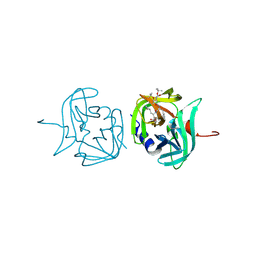 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 98 | | Descriptor: | 3C PROTEASE, N-(tert-butoxycarbonyl)-O-tert-butyl-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVD
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 83 | | Descriptor: | 3C PROTEASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
7QTK
 
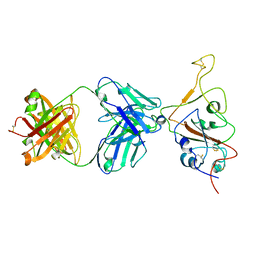 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, P2G3 Light Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
7QTI
 
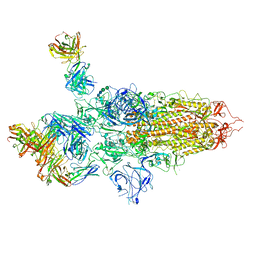 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
7QTJ
 
 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, P2G3 Light Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
7P40
 
 | | P5C3 is a potent fab neutralizer | | Descriptor: | Spike glycoprotein, Variable Heavy Chain P5C3 (VH), Variable Light Chain P5C3 (VL) | | Authors: | perez, L. | | Deposit date: | 2021-07-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
7PHG
 
 | | MaP OF P5C3RBD Interface | | Descriptor: | Heavy ChaIn variable, Light ChaIn, Surface glycoprotein | | Authors: | Perez, L. | | Deposit date: | 2021-08-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
3ZVE
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 84 | | Descriptor: | 3C PROTEASE, O-tert-butyl-N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVB
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 81 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-{[N-(TERT-BUTOXYCARBONYL)-L-PHENYLALANYL]AMINO}-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZV8
 
 | |
3ZVF
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 85 | | Descriptor: | 3C PROTEASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZV9
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 74 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVA
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 75 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVC
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 82 | | Descriptor: | 3C PROTEASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(3-TERT-BUTOXY-3-OXOPROPYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
4Y2A
 
 | | Crystal Structure of Coxsackie Virus B3 3D polymerase in complex with GPC-N114 inhibitor | | Descriptor: | 2,2'-[(4-chlorobenzene-1,2-diyl)bis(oxy)]bis(5-nitrobenzonitrile), 3D polymerase, GLYCEROL, ... | | Authors: | Vives-Adrian, L, Ferrer-Orta, C, Verdaguer, N. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
4Y3C
 
 | | I304V 3D polymerase mutant of EMCV | | Descriptor: | 3D polymerase, CHLORIDE ION, GLYCEROL | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
4Y34
 
 | | Crystal Structure of Coxsackievirus B3 3D polymerase in complex with GPC-N143 | | Descriptor: | 2,2'-[(4-fluorobenzene-1,2-diyl)bis(oxy)]bis(5-nitrobenzonitrile), 3D polymerase, GLYCEROL, ... | | Authors: | Vives-Adrian, L, Ferrer-Orta, C, Cerdaguer, N. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
