4PNH
 
 | |
1EKJ
 
 | |
4TN5
 
 | |
4EJ0
 
 | |
4WWX
 
 | | Crystal structure of the core RAG1/2 recombinase | | Descriptor: | V(D)J recombination-activating protein 1, V(D)J recombination-activating protein 2, ZINC ION | | Authors: | Kim, M.S, Lapkouski, M, Yang, W, Gellert, M. | | Deposit date: | 2014-11-12 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2001 Å) | | Cite: | Crystal structure of the V(D)J recombinase RAG1-RAG2.
Nature, 518, 2015
|
|
5ZE2
 
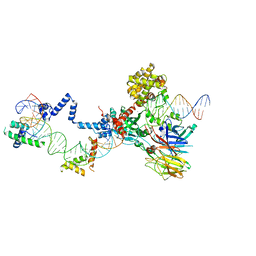 | | Hairpin Complex, RAG1/2-hairpin 12RSS/23RSS complex in 5mM Mn2+ for 2 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (31-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZE1
 
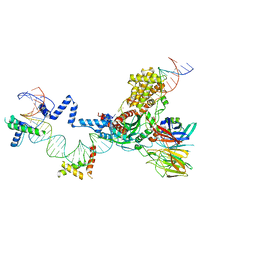 | | Hairpin Forming Complex, RAG1/2-Nicked 12RSS/23RSS complex in 2mM Mn2+ for 10 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA, HMGB1 A-B box, ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZDZ
 
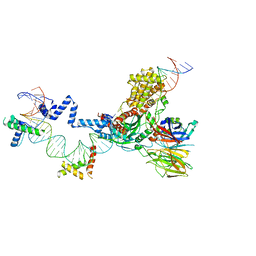 | | Hairpin Forming Complex, RAG1/2-Nicked 12RSS/23RSS complex in Ca2+ | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (30-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZE0
 
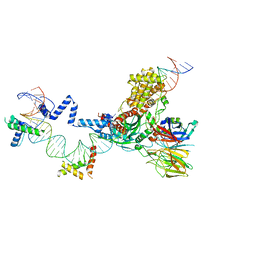 | | Hairpin Forming Complex, RAG1/2-Nicked(with Dideoxy) 12RSS/23RSS complex in Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (39-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
1F1W
 
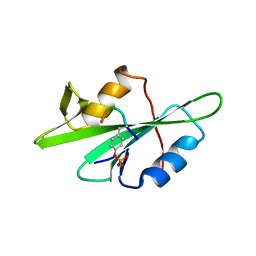 | | SRC SH2 THREF1TRP MUTANT COMPLEXED WITH THE PHOSPHOPEPTIDE S(PTR)VNVQN | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, S(PTR)VNVQN PHOSPHOPEPTIDE | | Authors: | Kimber, M.S, Nachman, J, Cunningham, A.M, Gish, G.D, Pawson, T, Pai, E.F. | | Deposit date: | 2000-05-20 | | Release date: | 2000-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specificity switching of the Src SH2 domain.
Mol.Cell, 5, 2000
|
|
1F2F
 
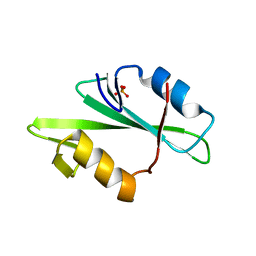 | | SRC SH2 THREF1TRP MUTANT | | Descriptor: | PHOSPHATE ION, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Kimber, M.S, Nachman, J, Cunningham, A.M, Gish, G.D, Pawson, T, Pai, E.F. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for specificity switching of the Src SH2 domain.
Mol.Cell, 5, 2000
|
|
3HLN
 
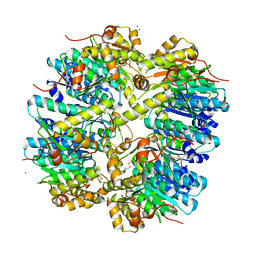 | | Crystal structure of ClpP A153C mutant with inter-heptamer disulfide bonds | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION | | Authors: | Kimber, M.S, Yu, A.Y.H, Borg, M, Chan, H.S, Houry, W.A. | | Deposit date: | 2009-05-27 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Theoretical Studies Indicate that the Cylindrical Protease ClpP Samples Extended and Compact Conformations.
Structure, 18, 2010
|
|
1U1Z
 
 | | The Structure of (3R)-hydroxyacyl-ACP dehydratase (FabZ) | | Descriptor: | (3R)-hydroxymyristoyl-[acyl carrier protein] dehydratase, SULFATE ION | | Authors: | Kimber, M.S, Martin, F, Lu, Y, Houston, S, Vedadi, M, Dharamsi, A, Fiebig, K.M, Schmid, M, Rock, C.O. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa
J.Biol.Chem., 279, 2004
|
|
7DR4
 
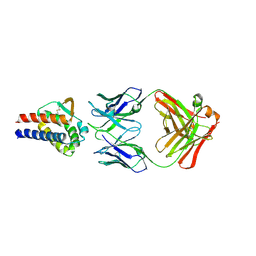 | | Complex of anti-human IL-2 antibody and human IL-2 | | Descriptor: | Interleukin-2, anti-human IL-2 antibody, mouse Ig G, ... | | Authors: | Kim, M.S, Kim, J.E. | | Deposit date: | 2020-12-25 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of human interleukin-2 in complex with TCB2, a new antibody-drug candidate with antitumor activity.
Oncoimmunology, 10, 2021
|
|
7SHG
 
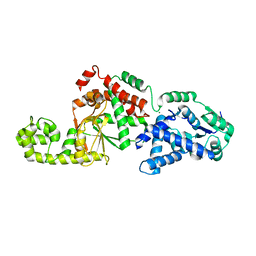 | |
4Y0C
 
 | | The structure of Arabidopsis ClpT2 | | Descriptor: | CHLORIDE ION, Clp protease-related protein At4g12060, chloroplastic, ... | | Authors: | Kimber, M.S, Schultz, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structures, Functions, and Interactions of ClpT1 and ClpT2 in the Clp Protease System of Arabidopsis Chloroplasts.
Plant Cell, 27, 2015
|
|
4Y0B
 
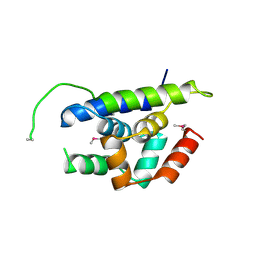 | | The structure of Arabidopsis ClpT1 | | Descriptor: | CHLORIDE ION, Double Clp-N motif protein | | Authors: | Kimber, M.S, Schultz, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-13 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures, Functions, and Interactions of ClpT1 and ClpT2 in the Clp Protease System of Arabidopsis Chloroplasts.
Plant Cell, 27, 2015
|
|
7SGY
 
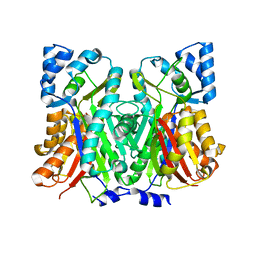 | |
4WEP
 
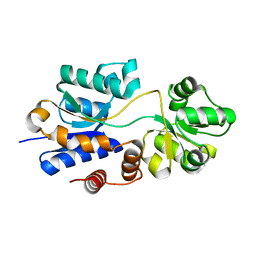 | | Apo YehZ from Escerichia coli | | Descriptor: | Putative osmoprotectant uptake system substrate-binding protein OsmF | | Authors: | Kimber, M.S, Lang, S, Mendoza, K, Wood, J.M. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | YehZYXW of Escherichia coli Is a Low-Affinity, Non-Osmoregulatory Betaine-Specific ABC Transporter.
Biochemistry, 54, 2015
|
|
6CIL
 
 | | PRE-REACTION COMPLEX, RAG1(E962Q)/2-INTACT/INTACT 12/23RSS COMPLEX IN MN2+ | | Descriptor: | High mobility group protein B1, Intact 12RSS substrate forward strand, Intact 12RSS substrate reverse strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CIM
 
 | | Pre-Reaction Complex, RAG1(E962Q)/2-nicked/intact 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 23RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CIK
 
 | | Pre-Reaction Complex, RAG1(E962Q)/2-intact/nicked 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 12RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6JFV
 
 | | The crystal structure of 2B-2B complex from keratins 5 and 14 (C367A mutant of K14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Kim, M.S, Lee, C.H, Coulombe, P.A, Leahy, D.J. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly.
Structure, 28, 2020
|
|
3NOJ
 
 | | The structure of HMG/CHA aldolase from the protocatechuate degradation pathway of Pseudomonas putida | | Descriptor: | 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate decarboxylase, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Kimber, M.S, Wang, W, Mazurkewich, S, Seah, S.Y.K. | | Deposit date: | 2010-06-25 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Kinetic Characterization of 4-Hydroxy-4-methyl-2-oxoglutarate/4-Carboxy-4-hydroxy-2-oxoadipate Aldolase, a Protocatechuate Degradation Enzyme Evolutionarily Convergent with the HpaI and DmpG Pyruvate Aldolases.
J.Biol.Chem., 285, 2010
|
|
6U4B
 
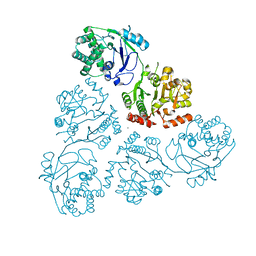 | | WbbM bifunctional glycosytransferase apo structure | | Descriptor: | MAGNESIUM ION, WbbM protein | | Authors: | Kimber, M.S, Mallette, E, Kamski-Hennekam, E.R, Gitalis, R. | | Deposit date: | 2019-08-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A bifunctional O-antigen polymerase structure reveals a new glycosyltransferase family.
Nat.Chem.Biol., 16, 2020
|
|
