3OXQ
 
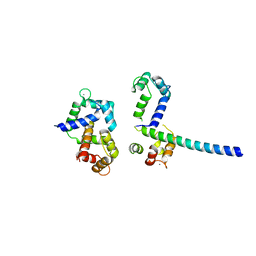 | | Crystal Structure of Ca2+/CaM-CaV1.2 pre-IQ/IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Kim, E.Y, Rumpf, C.H, Van Petegem, F, Arant, R, Findeisen, F, Cooley, E.S, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2010-09-21 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Multiple C-terminal tail Ca(2+)/CaMs regulate Ca(V)1.2 function but do not mediate channel dimerization.
Embo J., 29, 2010
|
|
2FTS
 
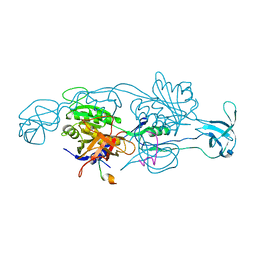 | |
2FU3
 
 | |
3DVE
 
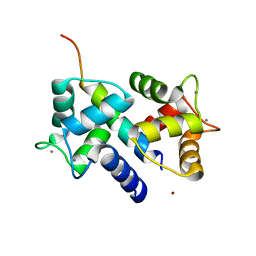 | | Crystal Structure of Ca2+/CaM-CaV2.2 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, NICKEL (II) ION, ... | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DVM
 
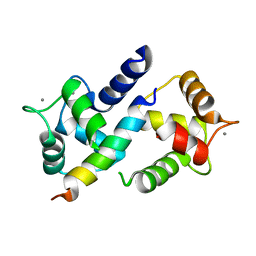 | | Crystal Structure of Ca2+/CaM-CaV2.1 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent P/Q-type calcium channel subunit alpha-1A | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DVJ
 
 | | Crystal Structure of Ca2+/CaM-CaV2.2 IQ domain (without cloning artifact, HM to TV) complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent N-type calcium channel subunit alpha-1B | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DVK
 
 | | Crystal Structure of Ca2+/CaM-CaV2.3 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent R-type calcium channel subunit alpha-1E | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
6W30
 
 | | Protein Tyrosine Phosphatase 1B Bound to Amorphadiene | | Descriptor: | Amorphadiene, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sarkar, A, Kim, E.Y, Hongdusit, A, Sankaran, B, Fox, J. | | Deposit date: | 2020-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Microbially Guided Discovery and Biosynthesis of Biologically Active Natural Products.
Acs Synth Biol, 10, 2021
|
|
7LFO
 
 | | Protein Tyrosine Phosphatase 1B | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Sarkar, A, Kim, E.Y, Hongdusit, A, Sankaran, B, Fox, J.M. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Microbially Guided Discovery and Biosynthesis of Biologically Active Natural Products.
Acs Synth Biol, 10, 2021
|
|
2HBA
 
 | | Crystal Structure of N-terminal Domain of Ribosomal Protein L9 (NTL9) K12M | | Descriptor: | 50S ribosomal protein L9, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Cho, J.-H, Kim, E.Y, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Energetically significant networks of coupled interactions within an unfolded protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2HVF
 
 | | Crystal Structure of N-terminal Domain of Ribosomal Protein L9 (NTL9), G34dA | | Descriptor: | 50S ribosomal protein L9, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Anil, B, Kim, E.Y, Cho, J.H, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-07-28 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Detecting and quantifying strain in protein folding
To be Published
|
|
2HBB
 
 | | Crystal Structure of the N-terminal Domain of Ribosomal Protein L9 (NTL9) | | Descriptor: | 50S ribosomal protein L9, ZINC ION | | Authors: | Cho, J.-H, Kim, E.Y, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Energetically significant networks of coupled interactions within an unfolded protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2DFK
 
 | | Crystal structure of the CDC42-Collybistin II complex | | Descriptor: | GLYCEROL, SULFATE ION, cell division cycle 42 isoform 1, ... | | Authors: | Xiang, S, Kim, E.Y, Connelly, J.J, Nassar, N, Kirsch, J, Winking, J, Schwarz, G, Schindelin, H. | | Deposit date: | 2006-03-02 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Cdc42 in Complex with Collybistin II, a Gephyrin-interacting Guanine Nucleotide Exchange Factor.
J.Mol.Biol., 359, 2006
|
|
8QU9
 
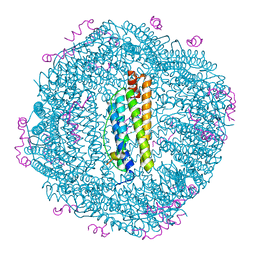 | | Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex | | Descriptor: | FE (III) ION, Ferritin heavy chain, NCOA4 (Nuclear Receptor Coactivator 4) | | Authors: | Hoelzgen, F, Klukin, E, Zalk, R, Shahar, A, Cohen-Schwartz, S, Frank, G.A. | | Deposit date: | 2023-10-15 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for the intracellular regulation of ferritin degradation.
Nat Commun, 15, 2024
|
|
7X0C
 
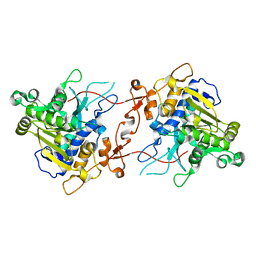 | | Crystal structure of phospholipase A1, AtDSEL | | Descriptor: | Phospholipase A1-IIgamma | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79909515 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
7X0D
 
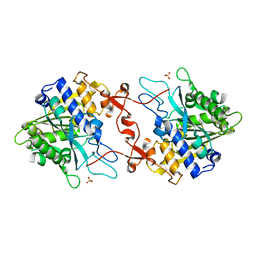 | | Crystal structure of phospholipase A1, CaPLA1 | | Descriptor: | Phospholipase A1, SULFATE ION | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39725161 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
