2V1O
 
 | | Crystal structure of N-terminal domain of acyl-CoA thioesterase 7 | | Descriptor: | COENZYME A, CYTOSOLIC ACYL COENZYME A THIOESTER HYDROLASE | | Authors: | Forwood, J.K, Thakur, A.S, Guncar, G, Marfori, M, Mouradov, D, Meng, W.N, Robinson, J, Huber, T, Kellie, S, Martin, J.L, Hume, D.A, Kobe, B. | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Recruitment of Tandem Hotdog Domains in Acyl-Coa Thioesterase 7 and its Role in Inflammation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2Y92
 
 | | Crystal structure of MAL adaptor protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TOLL/INTERLEUKIN-1 RECEPTOR DOMAIN-CONTAINING ADAPTER PROTEIN, | | Authors: | Valkov, E, Stamp, A, Martin, J.L, Kobe, B. | | Deposit date: | 2011-02-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of Toll-Like Receptor Adaptor Mal/Tirap Reveals the Molecular Basis for Signal Transduction and Disease Protection.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1WNH
 
 | | Crystal structure of mouse Latexin (tissue carboxypeptidase inhibitor) | | Descriptor: | Latexin | | Authors: | Aagaard, A, Listwan, P, Cowieson, N, Huber, T, Ravasi, T, Wells, C.A, Flanagan, J.U, Hume, D.A, Kobe, B, Martin, J.L. | | Deposit date: | 2004-08-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Inflammatory Role for the Mammalian Carboxypeptidase Inhibitor Latexin: Relationship to Cystatins and the Tumor Suppressor TIG1
Structure, 13, 2005
|
|
4DWN
 
 | | Crystal Structure of Human BinCARD CARD | | Descriptor: | Bcl10-interacting CARD protein, SULFATE ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2012-02-26 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | The structure of the caspase recruitment domain of BinCARD reveals that all three cysteines can be oxidized.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FH0
 
 | | Crystal Structure of Human BinCARD CARD, double mutant F16M/L66M SeMet form | | Descriptor: | Bcl10-interacting CARD protein, SULFATE ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2012-06-05 | | Release date: | 2013-02-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of the caspase recruitment domain of BinCARD reveals that all three cysteines can be oxidized.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3UX2
 
 | |
3UX3
 
 | | Crystal Structure of Domain-Swapped Fam96a minor dimer | | Descriptor: | ACETATE ION, MIP18 family protein FAM96A, ZINC ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2011-12-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mammalian DUF59 protein Fam96a forms two distinct types of domain-swapped dimer.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2Q2B
 
 | |
6WZW
 
 | | Ash1L SET domain in complex with AS-85 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Histone-lysine N-methyltransferase ASH1L, N-{[3-(3-carbamothioylphenyl)-1-{1-[(trifluoromethyl)sulfonyl]piperidin-4-yl}-1H-indol-6-yl]methyl}azetidine-3-carboxamide, ... | | Authors: | Li, H, Deng, J, Cierpicki, T, Grembecka, J. | | Deposit date: | 2020-05-14 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity.
Nat Commun, 12, 2021
|
|
6X0P
 
 | | Ash1L SET domain Q2265A mutant in complex with AS-5 | | Descriptor: | 3-[6-(aminomethyl)-1-(2-hydroxyethyl)-1H-indol-3-yl]benzene-1-carbothioamide, Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ... | | Authors: | Rogawski, D.S, Li, H, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2020-05-17 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity.
Nat Commun, 12, 2021
|
|
4QKO
 
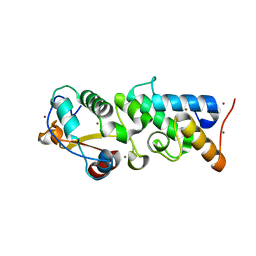 | | The Crystal Structure of the Pyocin S2 Nuclease Domain, Immunity Protein Complex at 1.8 Angstroms | | Descriptor: | BROMIDE ION, MAGNESIUM ION, Pyocin-S2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, C.J, Walker, D. | | Deposit date: | 2014-06-07 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into pyocin S2
To be Published
|
|
4QAY
 
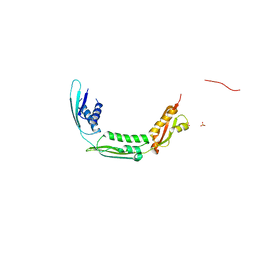 | |
4LED
 
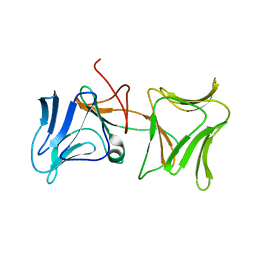 | | The Crystal Structure of Pyocin L1 bound to D-rhamnose at 2.37 Angstroms | | Descriptor: | Pyocin L1, alpha-D-rhamnopyranose | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4LEA
 
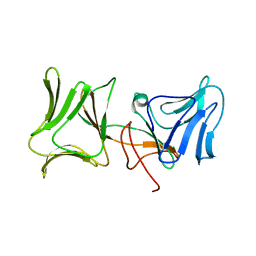 | | The Crystal Structure of Pyocin L1 bound to D-mannose at 2.55 Angstroms | | Descriptor: | Pyocin L1, beta-D-mannopyranose | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4LE7
 
 | | The Crystal Structure of Pyocin L1 at 2.09 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyocin L1 | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, R.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
2JPS
 
 | | NAB2 N-terminal domain | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2 | | Authors: | Grant, R, Marshall, N.J, Yang, J, Fasken, M, Kelly, S, Harreman, M.T, Neuhaus, D, Corbett, A.H, Stewart, M. | | Deposit date: | 2007-05-23 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2
J.Mol.Biol., 376, 2008
|
|
