6CPJ
 
 | | Solution structure of SH3 domain from Shank2 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 2 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
6CPK
 
 | | Solution structure of SH3 domain from Shank3 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
6CPI
 
 | | Solution structure of SH3 domain from Shank1 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
2ROA
 
 | | Solution structure of calcium bound soybean calmodulin isoform 4 N-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2RO9
 
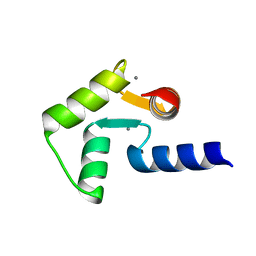 | | Solution structure of calcium bound soybean calmodulin isoform 1 C-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin-2 | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2ROB
 
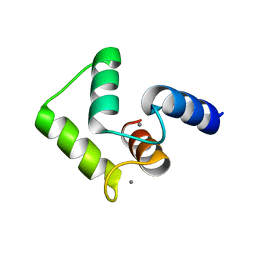 | | Solution structure of calcium bound soybean calmodulin isoform 4 C-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2RO8
 
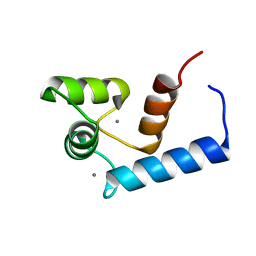 | | Solution structure of calcium bound soybean calmodulin isoform 1 N-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
1LKJ
 
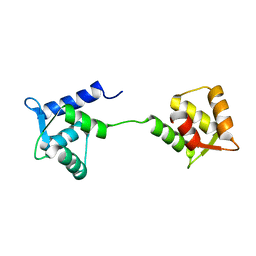 | | NMR Structure of Apo Calmodulin from Yeast Saccharomyces cerevisiae | | Descriptor: | Calmodulin | | Authors: | Ishida, H, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2002-04-25 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property.
Biochemistry, 41, 2002
|
|
5JOL
 
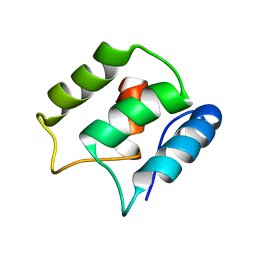 | | Calcium-free EF-hand domain of L-plastin | | Descriptor: | Plastin-2 | | Authors: | Ishida, H, Jensen, K.V, Woodman, A.G, Hyndman, M.E, Vogel, H.J. | | Deposit date: | 2016-05-02 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Calcium-Dependent Switch Helix of L-Plastin Regulates Actin Bundling.
Sci Rep, 7, 2017
|
|
5JOJ
 
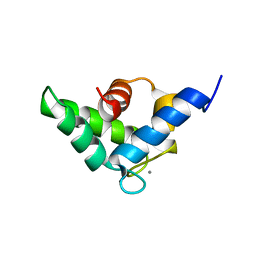 | | Calcium-loaded EF-hand domain of L-plastin | | Descriptor: | CALCIUM ION, Plastin-2 | | Authors: | Ishida, H, Jensen, K.V, Woodman, G.W, Hyndman, M.E, Vogel, H.J. | | Deposit date: | 2016-05-02 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Calcium-Dependent Switch Helix of L-Plastin Regulates Actin Bundling.
Sci Rep, 7, 2017
|
|
1F54
 
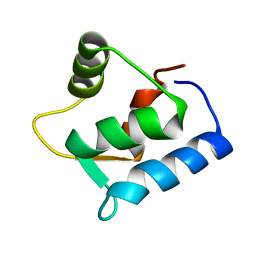 | | SOLUTION STRUCTURE OF THE APO N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
1F55
 
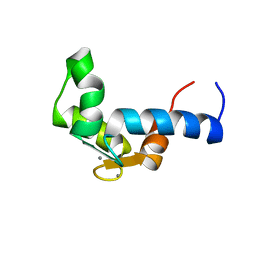 | | SOLUTION STRUCTURE OF THE CALCIUM BOUND N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
2L1W
 
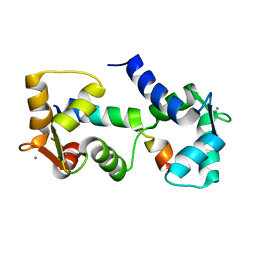 | |
2K3S
 
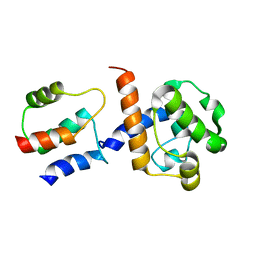 | | HADDOCK-derived structure of the CH-domain of the smoothelin-like 1 complexed with the C-domain of apocalmodulin | | Descriptor: | Calmodulin, Smoothelin-like protein 1 | | Authors: | Ishida, H, Borman, M.A, Ostrander, J, Vogel, H.J, MacDonald, J.A. | | Deposit date: | 2008-05-15 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the calponin homology (CH) domain from the smoothelin-like 1 protein: a unique apocalmodulin-binding mode and the possible role of the C-terminal type-2 CH-domain in smooth muscle relaxation.
J.Biol.Chem., 283, 2008
|
|
2JV9
 
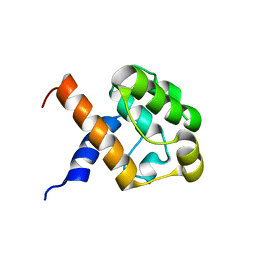 | |
2MQE
 
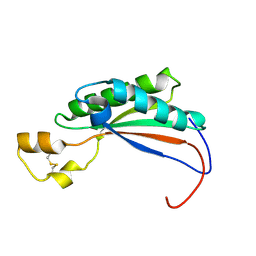 | |
2KN2
 
 | |
5ZLN
 
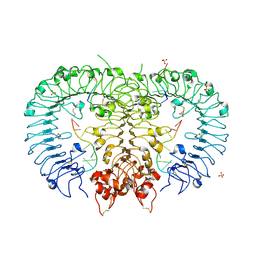 | | Crystal structure of mouse TLR9 in complex with two DNAs (CpG DNA and TCGCCA DNA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*AP*GP*GP*CP*GP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Ishida, H, Ohto, U, Shimizu, T. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for species-specific activation of mouse Toll-like receptor 9
FEBS Lett., 592, 2018
|
|
1NDH
 
 | |
2EX6
 
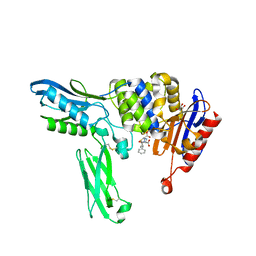 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
1IV4
 
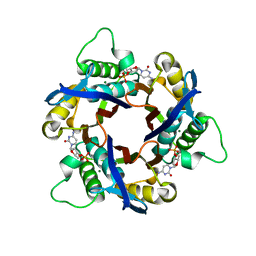 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form Substrate) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IV3
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form MG atoms) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IV2
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form CDP) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IV1
 
 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3A2F
 
 | |
