2EQX
 
 | | Solution structure of the BACK domain of Kelch repeat and BTB domain-containing protein 4 | | Descriptor: | Kelch repeat and BTB domain-containing protein 4 | | Authors: | Imai, M, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the BACK domain of Kelch repeat and BTB domain-containing protein 4
To be Published
|
|
2EPY
 
 | | Solution structure of the 10th C2H2 type zinc finger domain of Zinc finger protein 268 | | Descriptor: | ZINC ION, Zinc finger protein 268 | | Authors: | Imai, M, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 10th C2H2 type zinc finger domain of Zinc finger protein 268
To be Published
|
|
2EQW
 
 | | Solution structure of the 6th C2H2 type zinc finger domain of Zinc finger protein 484 | | Descriptor: | ZINC ION, Zinc finger protein 484 | | Authors: | Imai, M, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 6th C2H2 type zinc finger domain of Zinc finger protein 484
To be Published
|
|
2N9I
 
 | | Solution structure of reduced human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2N9J
 
 | | Solution structure of oxidized human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c.
Biochem.Biophys.Res.Commun., 469, 2016
|
|
3OG2
 
 | | Native crystal structure of Trichoderma reesei beta-galactosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Maksimainen, M, Rouvinen, J. | | Deposit date: | 2010-08-16 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of Trichoderma reesei beta-galactosidase reveal conformational changes in the active site
J.Struct.Biol., 174, 2011
|
|
3OGV
 
 | | Complex structure of beta-galactosidase from Trichoderma reesei with PETG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-phenylethyl 1-thio-beta-D-galactopyranoside, ... | | Authors: | Maksimainen, M, Rouvinen, J. | | Deposit date: | 2010-08-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of Trichoderma reesei beta-galactosidase reveal conformational changes in the active site
J.Struct.Biol., 174, 2011
|
|
3OGR
 
 | | Complex structure of beta-galactosidase from Trichoderma reesei with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Maksimainen, M, Rouvinen, J. | | Deposit date: | 2010-08-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of Trichoderma reesei beta-galactosidase reveal conformational changes in the active site
J.Struct.Biol., 174, 2011
|
|
3OGS
 
 | | Complex structure of beta-galactosidase from Trichoderma reesei with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maksimainen, M, Rouvinen, J. | | Deposit date: | 2010-08-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Trichoderma reesei beta-galactosidase reveal conformational changes in the active site
J.Struct.Biol., 174, 2011
|
|
3TTS
 
 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus | | Descriptor: | Beta-galactosidase, ZINC ION | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
3TTY
 
 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus in complex with galactose | | Descriptor: | Beta-galactosidase, ZINC ION, alpha-D-galactopyranose | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
4IUG
 
 | | Crystal structure of beta-galactosidase from Aspergillus oryzae in complex with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-6-O-phosphono-beta-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, ... | | Authors: | Maksimainen, M, Rouvinen, J. | | Deposit date: | 2013-01-21 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of acidic beta-galactosidase from Aspergillus oryzae.
Int.J.Biol.Macromol., 60C, 2013
|
|
8DOX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8DPR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with inhibitor TKB-248 | | Descriptor: | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-16 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
3KHW
 
 | |
3L56
 
 | | Crystal structure of the large c-terminal domain of polymerase basic protein 2 from influenza virus a/viet nam/1203/2004 (h5n1) | | Descriptor: | Polymerase PB2 | | Authors: | Staker, B.L, Edwards, T, Eric, S, Raymond, A, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-12-21 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biological and structural characterization of a host-adapting amino acid in influenza virus.
Plos Pathog., 6, 2010
|
|
3KC6
 
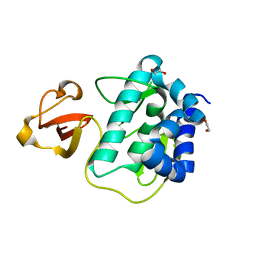 | |
6K4H
 
 | | Crystal structure of the PI5P4Kbeta-AMPPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Senda, M, Senda, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 2022
|
|
6K4G
 
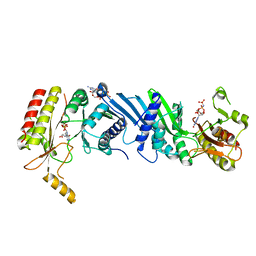 | | Crystal structure of the PI5P4Kbeta-GMPPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Senda, M, Senda, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 2022
|
|
7Y6L
 
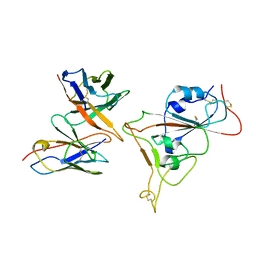 | |
7Y6N
 
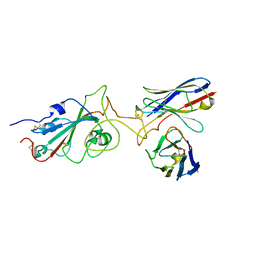 | |
7X93
 
 | |
7X95
 
 | |
7X94
 
 | |
7X96
 
 | |
