9BDQ
 
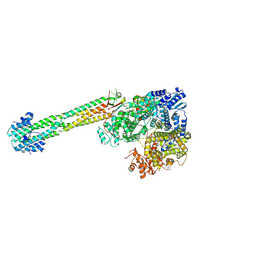 | | The structure of NiV L-P complex | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Hu, S, Yang, P, Yu, Z, Abraham, J. | | Deposit date: | 2024-04-12 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Structural and functional analysis of the Nipah virus polymerase complex.
Cell, 188, 2025
|
|
3WLW
 
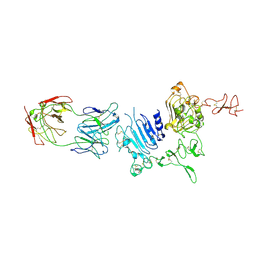 | | Molecular Architecture of the ErbB2 Extracellular Domain Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody H Chain, ... | | Authors: | Hu, S, Lou, Z.Y, Guo, Y.J. | | Deposit date: | 2013-11-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.088 Å) | | Cite: | Molecular architecture of the ErbB2 extracellular domain homodimer.
Oncotarget, 6, 2015
|
|
3WD5
 
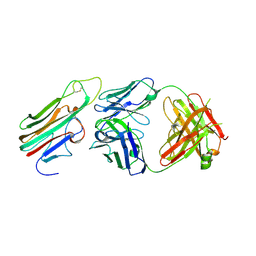 | | Crystal structure of TNFalpha in complex with Adalimumab Fab fragment | | Descriptor: | Adalimumab Heavy Chain, Adalimumab Light Chain, Tumor necrosis factor | | Authors: | Hu, S, Liang, S.Y, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2013-06-06 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor alpha-associated diseases from a molecular view
J.Biol.Chem., 288, 2013
|
|
6J56
 
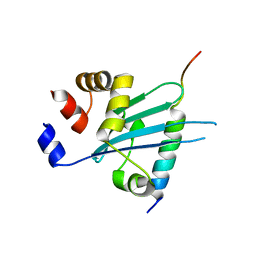 | | Crystal structure of Myosin VI CBD in complex with Tom1 MBM | | Descriptor: | Peptide from Target of Myb protein 1, Unconventional myosin-VI | | Authors: | Hu, S, Pan, L. | | Deposit date: | 2019-01-10 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structure of Myosin VI/Tom1 complex reveals a cargo recognition mode of Myosin VI for tethering.
Nat Commun, 10, 2019
|
|
7F5P
 
 | | The crystal structure of VyPAL2-C214A, a dead mutant of VyPAL2 peptide asparaginyl ligase in form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide Asparaginyl Ligases, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5J
 
 | | The crystal structure of VyPAL2-I244V, a more efficient mutant of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5Q
 
 | | The crystal structure of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7FA0
 
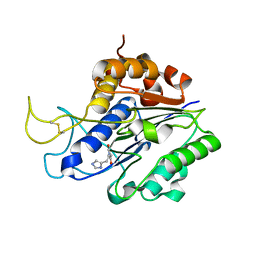 | | The crystal structure of VyPAL2-C214A, a dead mutant of VyPAL2 peptide asparaginyl ligase in form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide Asparaginyl Ligases, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-07-05 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
4WHA
 
 | | Lipoxygenase-1 (soybean) L546A/L754A mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (III) ION, ... | | Authors: | Scouras, A.D, Carr, C.A.M, Hu, S, Klinman, J.P. | | Deposit date: | 2014-09-21 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extremely elevated room-temperature kinetic isotope effects quantify the critical role of barrier width in enzymatic C-H activation.
J.Am.Chem.Soc., 136, 2014
|
|
3W94
 
 | |
5YT6
 
 | |
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
3C8C
 
 | | Crystal structure of Mcp_N and cache domains of methyl-accepting chemotaxis protein from Vibrio cholerae | | Descriptor: | ALANINE, MAGNESIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Hu, S, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-11 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Mcp_N and cache N-terminal domains of methyl-accepting chemotaxis protein from Vibrio cholerae.
To be Published
|
|
3CAX
 
 | | Crystal structure of uncharacterized protein PF0695 | | Descriptor: | Uncharacterized protein PF0695 | | Authors: | Ramagopal, U.A, Hu, S, Toro, R, Gilmore, M, Bain, K, Meyer, A.J, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of uncharacterized protein PF0695.
To be Published
|
|
3CMG
 
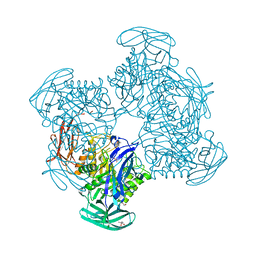 | | Crystal structure of putative beta-galactosidase from Bacteroides fragilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Ramagopal, U.A, Rutter, M, Toro, R, Hu, S, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-21 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative beta-galactosidase from Bacteroides fragilis.
To be published
|
|
2RDM
 
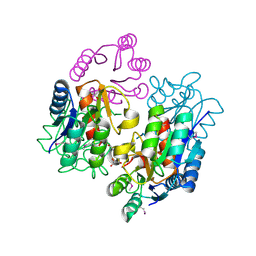 | | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Yan, Q, Zhan, C, Toro, R, Meyer, A.J, Gilmore, M, Hu, S, Groshong, C, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419.
To be Published
|
|
3DOU
 
 | | Crystal structure of methyltransferase involved in cell division from thermoplasma volcanicum gss1 | | Descriptor: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYLMETHIONINE | | Authors: | Patskovsky, Y, Ozyurt, S, Dickey, M, Hu, S, Bain, K, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-06 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Methyltransferase Involved in Cell Division from Thermoplasma Volcanicum
To be Published
|
|
3EMU
 
 | | Crystal structure of a leucine rich repeat and phosphatase domain containing protein from Entamoeba histolytica | | Descriptor: | SULFATE ION, leucine rich repeat and phosphatase domain containing protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a leucine rich repeat and phosphatase domain containing protein from Entamoeba histolytica
To be Published
|
|
3EEZ
 
 | | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Silicibacter pomeroyi | | Descriptor: | putative Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-07 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Silicibacter pomeroyi
To be Published
|
|
3DBY
 
 | | Crystal structure of uncharacterized protein from Bacillus cereus G9241 (CSAP Target) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, uncharacterized protein | | Authors: | Ramagopal, U.A, Bonanno, J.B, Ozyurt, S, Freeman, J, Wasserman, S, Hu, S, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uncharacterized protein from Bacillus cereus G9241
To be published
|
|
3E03
 
 | | Crystal structure of a putative dehydrogenase from Xanthomonas campestris | | Descriptor: | CALCIUM ION, Short chain dehydrogenase | | Authors: | Sampathkumar, P, Wasserman, S, Rutter, M, Hu, S, Bain, K, Rodgers, L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of a putative dehydrogenase from Xanthomonas campestris
To be Published
|
|
3ICJ
 
 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus furiosus | | Descriptor: | ZINC ION, uncharacterized metal-dependent hydrolase | | Authors: | Bonanno, J.B, Patskovsky, Y, Freeman, J, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Furiosus
To be Published
|
|
3E8V
 
 | | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis | | Descriptor: | Possible transglutaminase-family protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis
To be Published
|
|
3E2V
 
 | | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae | | Descriptor: | 3'-5'-exonuclease, GLYCEROL, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae
To be Published
|
|
3E9N
 
 | | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum | | Descriptor: | PUTATIVE SHORT-CHAIN DEHYDROGENASE/REDUCTASE | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum
To be Published
|
|
