7KSE
 
 | | Crystal structure of Prototype Foamy Virus Protease-Reverse Transcriptase CSH mutant (selenomethionine-labeled) | | Descriptor: | CALCIUM ION, Peptidase A9/Reverse transcriptase/RNase H | | Authors: | Harrison, J.J.E.K, Das, K, Ruiz, F.X, Arnold, E. | | Deposit date: | 2020-11-21 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a Retroviral Polyprotein: Prototype Foamy Virus Protease-Reverse Transcriptase (PR-RT).
Viruses, 13, 2021
|
|
7KSF
 
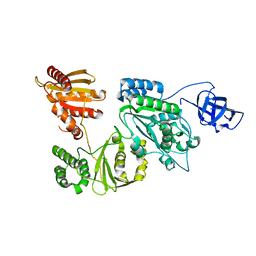 | | Crystal structure of Prototype Foamy Virus Protease-Reverse Transcriptase (native) | | Descriptor: | CALCIUM ION, Protease/Reverse transcriptase/ribonuclease H | | Authors: | Harrison, J.J.E.K, Das, K, Ruiz, F.X, Arnold, E. | | Deposit date: | 2020-11-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Retroviral Polyprotein: Prototype Foamy Virus Protease-Reverse Transcriptase (PR-RT).
Viruses, 13, 2021
|
|
7SEP
 
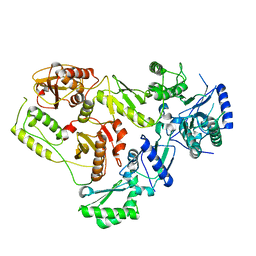 | |
7SJX
 
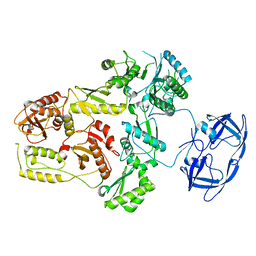 | | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | | Descriptor: | Gag-Pol polyprotein | | Authors: | Lyumkis, D, Passos, D, Arnold, E, Harrison, J.J.E.K, Ruiz, F.X. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Sci Adv, 8, 2022
|
|
6WJ9
 
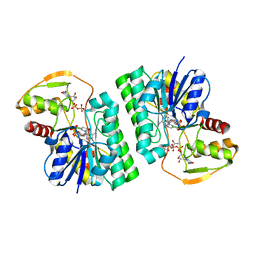 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
7L30
 
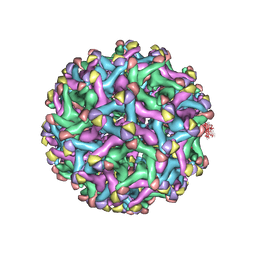 | | Binjari virus (BinJV) | | Descriptor: | Envelope protein E, prM protein | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-12-17 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The structure of an infectious immature flavivirus redefines viral architecture and maturation.
Sci Adv, 7, 2021
|
|
5T10
 
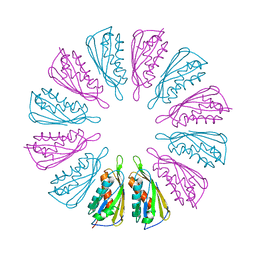 | |
5T0Z
 
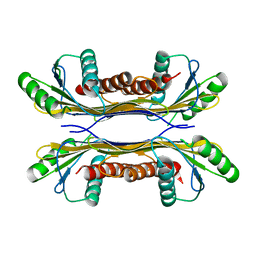 | | PelC from Geobacter metallireducens | | Descriptor: | Lipoprotein, putative | | Authors: | Marmont, L.S, Howell, P.L. | | Deposit date: | 2016-08-17 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Oligomeric lipoprotein PelC guides Pel polysaccharide export across the outer membrane of Pseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T11
 
 | |
6WJB
 
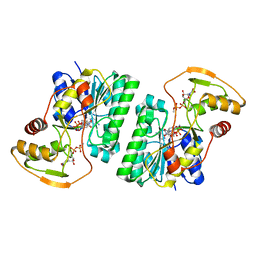 | | UDP-GlcNAc C4-epimerase from Pseudomonas protegens in complex with NAD and UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Pfoh, R, Robinson, H, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
5CXR
 
 | |
5CYQ
 
 | | HIV-1 reverse transcriptase complexed with 4-bromopyrazole | | Descriptor: | 4-bromo-1H-pyrazole, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, BROMIDE ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Rapid experimental SAD phasing and hot-spot identification with halogenated fragments.
IUCrJ, 3, 2016
|
|
6WJA
 
 | |
5WFT
 
 | | PelB 319-436 from Pseudomonas aeruginosa PAO1 | | Descriptor: | PelB | | Authors: | Marmont, L.S, Howell, P.L. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa.
J. Biol. Chem., 292, 2017
|
|
5CYM
 
 | | HIV-1 reverse transcriptase complexed with 4-iodopyrazole | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Iucrj, 3, 2016
|
|
5CW1
 
 | | Proteinase K complexed with 4-iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, IODIDE ION, Proteinase K, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Iucrj, 3, 2016
|
|
