8DQV
 
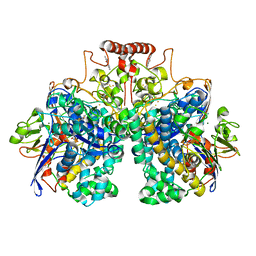 | | The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.52 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UUR
 
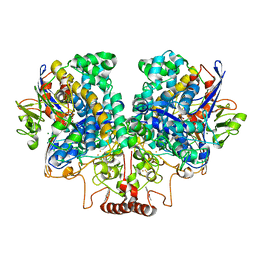 | | The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, HYDROXIDE ION, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UUS
 
 | | The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UTD
 
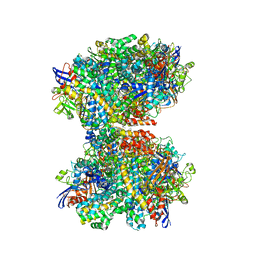 | | The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-26 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
6UW1
 
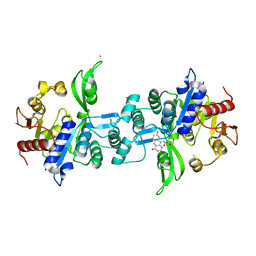 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UVX
 
 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Apo state | | Descriptor: | CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW5
 
 | | The crystal structure of FbiA from Mycobacterium smegmatis, GDP and Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW7
 
 | | The crystal structure of FbiA from Mycobacterium smegmatis, Dehydro-F420-0 bound form | | Descriptor: | 2-[oxidanyl-[(2~{R},3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-[2,4,8-tris(oxidanylidene)-1,9-dihydropyrimido[4,5-b]quinolin-10-yl]pentoxy]phosphoryl]oxyprop-2-enoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Izore, T, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW3
 
 | | The crystal structure of FbiA from Mycobacterium Smegmatis, GDP Bound form | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
1U16
 
 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) in complex with sulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Delta crystallin I, ... | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
1U15
 
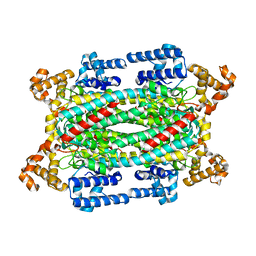 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) | | Descriptor: | Delta crystallin I | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
6ECI
 
 | | Structure of the FAD binding protein MSMEG_5243 from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pyridoxamine 5'-phosphate oxidase-related, ... | | Authors: | Ahmed, F.H, Antoney, J, Carr, P.D, Jackson, C.J. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | FAD-sequestering proteins protect mycobacteria against hypoxic and oxidative stress.
J. Biol. Chem., 294, 2019
|
|
5BNC
 
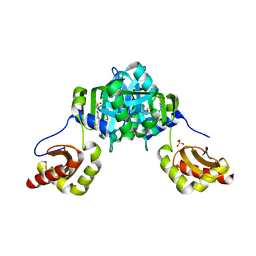 | | Structure of heme binding protein MSMEG_6519 from Mycobacterium smegmatis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ahmed, F.H, Carr, P.D, Jackson, C.J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
8UEM
 
 | |
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
8OKH
 
 | |
6BWG
 
 | |
6BWH
 
 | |
6OFT
 
 | |
6OFS
 
 | |
6OFR
 
 | |
4ZKY
 
 | | Structure of F420 binding protein, MSMEG_6526, from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, IODIDE ION, Pyridoxamine 5-phosphate oxidase, ... | | Authors: | Lee, B.M, Carr, P.D, Ahmed, F.H, Jackson, C.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
4Y9I
 
 | |
4YBN
 
 | | Structure of the FAD and Heme binding protein msmeg_4975 from Mycobacterium smegmatis | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-nucleotide-binding protein, ... | | Authors: | Ahmed, F.H, Carr, P.D, Jackson, C.J. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
6V4V
 
 | |
