4AAG
 
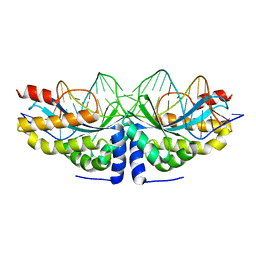 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target in presence of Ca at the active site (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*CP *GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', CALCIUM ION, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
3OEZ
 
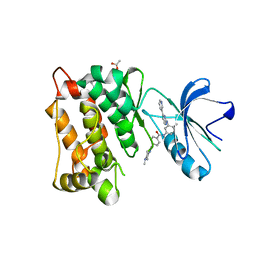 | | crystal structure of the L317I mutant of the chicken c-Src tyrosine kinase domain complexed with imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, ACETATE ION, GLYCEROL, ... | | Authors: | Boubeva, R, Pernot, L, Perozzo, R, Scapozza, L. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | a single amino-acid dictates the dynamics of the switch between active and inactive C-Src conformation
To be Published
|
|
3OF0
 
 | |
3QLG
 
 | |
3QLF
 
 | |
