3H0C
 
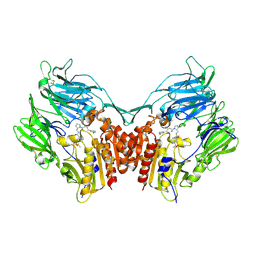 | | Crystal Structure of Human Dipeptidyl Peptidase IV (CD26) in Complex with a Reversed Amide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-({(2S)-1-[(3R)-3-amino-4-(3-chlorophenyl)butanoyl]pyrrolidin-2-yl}methyl)-3-(methylsulfonyl)benzamide | | Authors: | Nordhoff, S, Cerezo-Galvez, S, Deppe, H, Hill, O, Lopez-Canet, M, Rummey, C, Thiemann, M, Matassa, V.G, Edwards, P.J, Feurer, A. | | Deposit date: | 2009-04-09 | | Release date: | 2009-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of b-homophenylalanine based pyrrolidin-2-ylmethyl amides and sulfonamides as highly potent and selective inhibitors of dipeptidyl peptidase IV
To be Published
|
|
2BUC
 
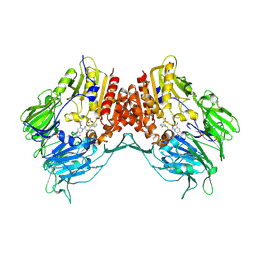 | | Crystal Structure Of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Tetrahydroisoquinoline Inhibitor | | Descriptor: | (S)-2-[(R)-3-AMINO-4-(2-FLUORO-PHENYL)-BUTYRYL]-1,2,3,4-TETRAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID AMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nordhoff, S, Cerezo-Galvez, S, Feurer, A, Hill, O, Matassa, V.G, Metz, G, Rummey, C, Thiemann, M, Edwards, P.J. | | Deposit date: | 2005-06-09 | | Release date: | 2006-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The reversed binding of beta-phenethylamine inhibitors of DPP-IV: X-ray structures and properties of novel fragment and elaborated inhibitors.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
2BUA
 
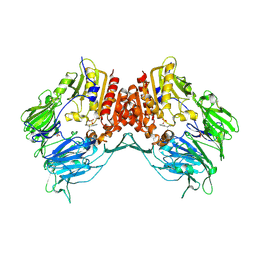 | | Crystal Structure Of Porcine Dipeptidyl Peptidase IV (Cd26) in Complex With a Low Molecular Weight Inhibitor. | | Descriptor: | 1-METHYLAMINE-1-BENZYL-CYCLOPENTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nordhoff, S, Cerezo-Galvez, S, Feurer, A, Hill, O, Matassa, V.G, Metz, G, Rummey, C, Thiemann, M, Edwards, P.J. | | Deposit date: | 2005-06-09 | | Release date: | 2006-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The reversed binding of beta-phenethylamine inhibitors of DPP-IV: X-ray structures and properties of novel fragment and elaborated inhibitors.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
2BUB
 
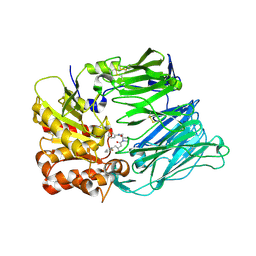 | | Crystal Structure Of Human Dipeptidyl Peptidase IV (CD26) in Complex with a Reversed Amide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIPEPTIDYL PEPTIDASE 4, N-({(2S)-1-[(3R)-3-AMINO-4-(2-FLUOROPHENYL)BUTANOYL]PYRROLIDIN-2-YL}METHYL)BENZAMIDE | | Authors: | Nordhoff, S, Cerezo-Galvez, S, Feurer, A, Hill, O, Matassa, V.G, Metz, G, Rummey, C, Thiemann, M, Edwards, P.J. | | Deposit date: | 2005-06-09 | | Release date: | 2006-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The reversed binding of beta-phenethylamine inhibitors of DPP-IV: X-ray structures and properties of novel fragment and elaborated inhibitors.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
8PWR
 
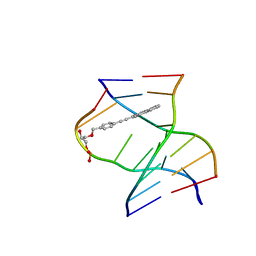 | | TINA-conjugated antiparallel DNA triplex | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*GP*GP*TP*GP*(J32)P*GP*T)-3') | | Authors: | Garavis, M, Edwards, P.J.B, Serrano-Chacon, I, Doluca, O, Filichev, V.V, Gonzalez, C. | | Deposit date: | 2023-07-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-27 | | Method: | SOLUTION NMR | | Cite: | Understanding intercalative modulation of G-rich sequence folding: solution structure of a TINA-conjugated antiparallel DNA triplex.
Nucleic Acids Res., 52, 2024
|
|
6U1L
 
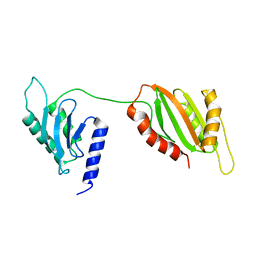 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
6U1O
 
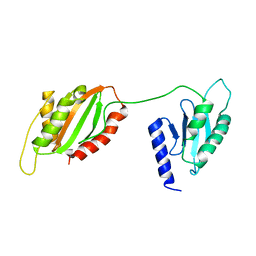 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
2KXG
 
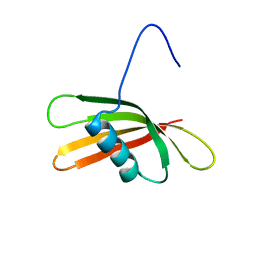 | | The solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) | | Descriptor: | Aspartic protease inhibitor | | Authors: | Headey, S.J, Macaskill, U.K, Wright, M, Claridge, J.K, Edwards, P.J.B, Farley, P.C, Christeller, J.T, Laing, W.A, Pascal, S.M. | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) and mutational analysis of pepsin inhibition.
J.Biol.Chem., 285, 2010
|
|
8DFZ
 
 | |
6BQI
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 2020
|
|
2KUY
 
 | | Structure of Glycocin F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prebacteriocin glycocin F | | Authors: | Venugopal, H, Edwards, P, Schwalbe, M, Claridge, J, Stepper, J, Patchett, M, Loo, T, Libich, D, Norris, G, Pascal, S. | | Deposit date: | 2010-03-01 | | Release date: | 2011-05-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural, dynamic, and chemical characterization of a novel s-glycosylated bacteriocin.
Biochemistry, 50, 2011
|
|
6B34
 
 | |
6B35
 
 | |
6ANM
 
 | |
6ANN
 
 | |
