1SL7
 
 | | Crystal structure of calcium-loaded apo-obelin from Obelia longissima | | Descriptor: | CALCIUM ION, Obelin | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
1S36
 
 | | Crystal structure of a Ca2+-discharged photoprotein: Implications for the mechanisms of the calcium trigger and the bioluminescence | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, ... | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.-J, Lee, J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-01-12 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a Ca2+-discharged photoprotein: implications for mechanisms of the calcium trigger and bioluminescence
J.Biol.Chem., 279, 2004
|
|
1SL9
 
 | | Obelin from Obelia longissima | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Deng, L, Markova, S, Vysotski, E, Liu, Z.-J, Lee, J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Obelin from Obelia longissima
To be Published
|
|
1SL8
 
 | | Calcium-loaded apo-aequorin from Aequorea victoria | | Descriptor: | Aequorin 1, CALCIUM ION | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
1JF0
 
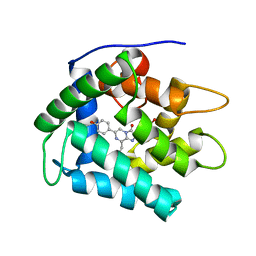 | | The Crystal Structure of Obelin from Obelia geniculata at 1.82 A Resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Deng, L, Vysotski, E, Liu, Z.-J, Markova, S, Lee, J, Rose, J, Wang, B.-C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Obelin from Obelia geniculata at 1.82 Angstrom Resolution
To be Published
|
|
3M18
 
 | |
3M19
 
 | |
2IAM
 
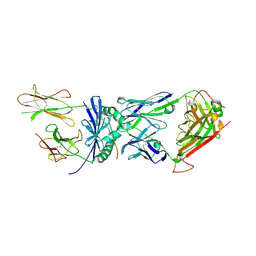 | | Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR | | Descriptor: | 15-mer peptide from Triosephosphate isomerase, CD4+ T cell receptor E8 alpha chain, CD4+ T cell receptor E8 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Mariuzza, R.A. | | Deposit date: | 2006-09-08 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor
Nat.Immunol., 8, 2007
|
|
2IAN
 
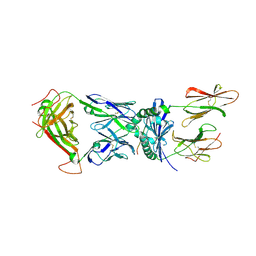 | | Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR | | Descriptor: | 15-mer peptide from Triosephosphate isomerase, CD4+ T cell receptor E8 alpha chain, CD4+ T cell receptor E8 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Mariuzza, R.A. | | Deposit date: | 2006-09-08 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor
Nat.Immunol., 8, 2007
|
|
2IAL
 
 | | Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR | | Descriptor: | CD4+ T cell receptor E8 alpha chain, CD4+ T cell receptor E8 beta chain | | Authors: | Deng, L, Langley, R.J, Mariuzza, R.A. | | Deposit date: | 2006-09-08 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor
Nat.Immunol., 8, 2007
|
|
1PRY
 
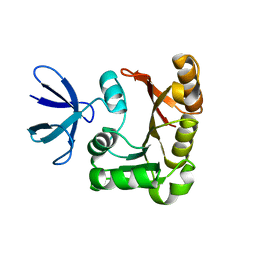 | | Structure Determination of Fibrillarin Homologue From Hyperthermophilic Archaeon Pyrococcus furiosus (Pfu-65527) | | Descriptor: | Fibrillarin-like pre-rRNA processing protein | | Authors: | Deng, L, Starostina, N.G, Liu, Z.-J, Rose, J.P, Terns, R.M, Terns, M.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-06-20 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure determination of fibrillarin from the hyperthermophilic archaeon Pyrococcus furiosus
Biochem.Biophys.Res.Commun., 315, 2004
|
|
1YWO
 
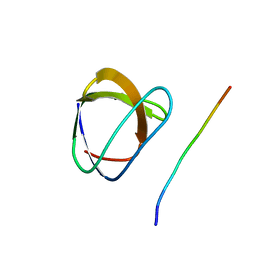 | | Phospholipase Cgamma1 SH3 in complex with a SLP-76 motif | | Descriptor: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1, Lymphocyte cytosolic protein 2 | | Authors: | Deng, L, Velikovsky, C.A, Swaminathan, C.P, Cho, S, Mariuzza, R.A. | | Deposit date: | 2005-02-18 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for Recognition of the T Cell Adaptor Protein SLP-76 by the SH3 Domain of Phospholipase Cgamma1
J.Mol.Biol., 352, 2005
|
|
1YWP
 
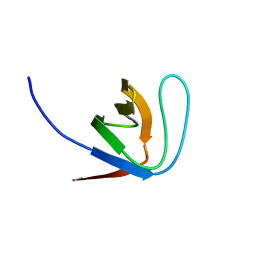 | | Phospholipase Cgamma1 SH3 | | Descriptor: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 | | Authors: | Deng, L, Velikovsky, C.A, Swaminathan, C.P, Cho, S, Mariuzza, R.A. | | Deposit date: | 2005-02-18 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Recognition of the T Cell Adaptor Protein SLP-76 by the SH3 Domain of Phospholipase Cgamma1
J.Mol.Biol., 352, 2005
|
|
3C8J
 
 | |
3C8K
 
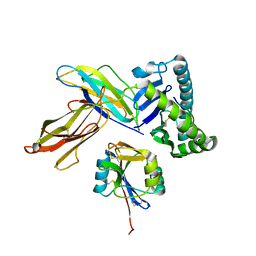 | | The crystal structure of Ly49C bound to H-2Kb | | Descriptor: | H-2 class I histocompatibility antigen, K-B alpha chain, Natural killer cell receptor Ly-49C, ... | | Authors: | Deng, L, Mariuzza, R.A. | | Deposit date: | 2008-02-12 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular architecture of the major histocompatibility complex class I-binding site of Ly49 natural killer cell receptors.
J.Biol.Chem., 283, 2008
|
|
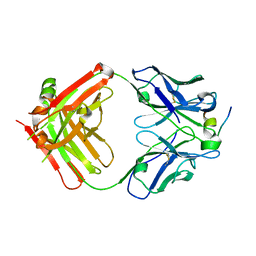
4HZL
 
 | |
7LKI
 
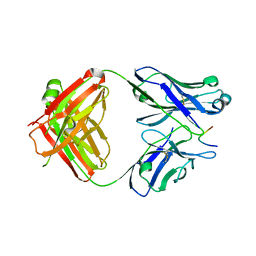 | |
4Q0X
 
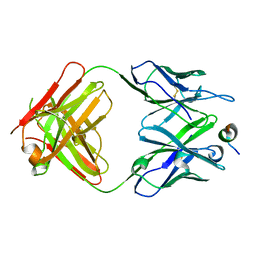 | |
4E42
 
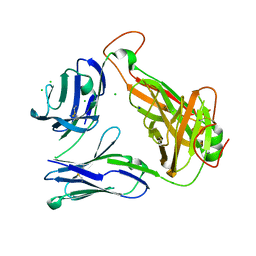 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | CHLORIDE ION, NITRATE ION, SODIUM ION, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
4E41
 
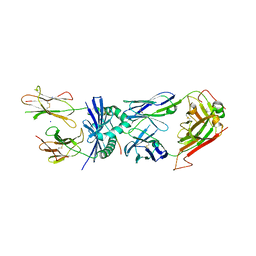 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
2PTU
 
 | | Structure of NK cell receptor 2B4 (CD244) | | Descriptor: | Natural killer cell receptor 2B4 | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
2PTV
 
 | | Structure of NK cell receptor ligand CD48 | | Descriptor: | CD48 antigen | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
3RAS
 
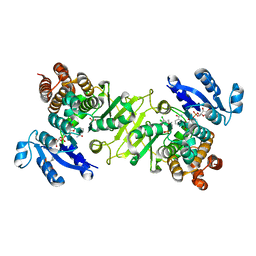 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with a lipophilic phosphonate inhibitor | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-(N-HYDROXYACETAMIDO)-1-(3,4-DICHLOROPHENYL)PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Diao, J, Deng, L, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase by lipophilic phosphonates: SAR, QSAR, and crystallographic studies.
J.Med.Chem., 54, 2011
|
|
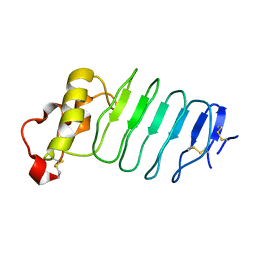
3G39
 
 | |
2PTT
 
 | |
