2JV7
 
 | |
1SA8
 
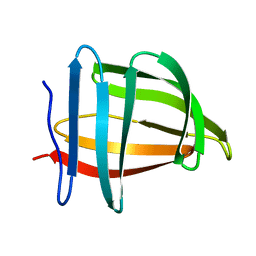 | |
2MM3
 
 | | Solution NMR structure of the ternary complex of human ileal bile acid-binding protein with glycocholate and glycochenodeoxycholate | | Descriptor: | GLYCOCHENODEOXYCHOLIC ACID, GLYCOCHOLIC ACID, Gastrotropin | | Authors: | Horvath, G, Egyed, O, Bencsura, A, Simon, A, Tochtrop, G.P, DeKoster, G.T, Covey, D.F, Cistola, D.P, Toke, O. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of ligand binding in the ternary complex of human ileal bile acid binding protein with glycocholate and glycochenodeoxycholate obtained from solution NMR
FEBS J., 283, 2016
|
|
6F4G
 
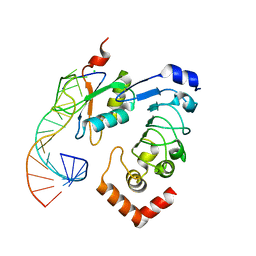 | | 'Crystal structure of the Drosophila melanogaster SNF/U2A'/U2-SL4 complex | | Descriptor: | CHLORIDE ION, Probable U2 small nuclear ribonucleoprotein A', SULFATE ION, ... | | Authors: | Weber, G, DeKoster, G, Holton, N, Hall, K.B, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6F4J
 
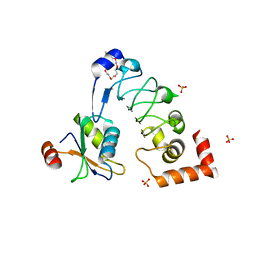 | | Crystal structure of Drosophila melanogaster SNF/U2A' complex | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Probable U2 small nuclear ribonucleoprotein A', SULFATE ION, ... | | Authors: | Weber, G, DeKoster, G, Holton, N, Hall, K.B, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6F4I
 
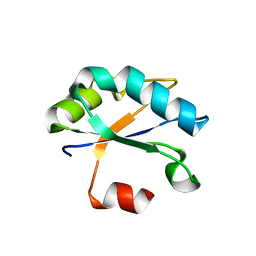 | | Crystal structure of Drosophila melanogaster SNF | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Weber, G, Holton, N, Hall, K.B, DeKoster, G, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6F4H
 
 | | Crystal structure of the Drosophila melanogaster SNF/U1-SL2 complex | | Descriptor: | CITRATE ANION, U1 small nuclear ribonucleoprotein A, U1-SL2 | | Authors: | Weber, G, Holton, N, Hall, K.B, DeKoster, G, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6V5T
 
 | |
6V64
 
 | | Crystal structure of human thrombin bound to ppack with tryptophans replaced by 5-F-tryptophan | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Ruben, E.A, Chen, Z, Di Cera, E. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.
J.Biol.Chem., 295, 2020
|
|
4NUV
 
 | |
4NUU
 
 | |
1T8V
 
 | |
