1VZV
 
 | | STRUCTURE OF VARICELLA-ZOSTER VIRUS PROTEASE | | Descriptor: | VARICELLA-ZOSTER VIRUS PROTEASE | | Authors: | Qiu, X, Jason, C.A, Culp, J.S, Richardson, S.B, Debouck, C, Smith, W.W, Abdel-Meguid, S.S. | | Deposit date: | 1997-02-10 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of varicella-zoster virus protease.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1HEF
 
 | | The crystal structures at 2.2 angstroms resolution of hydroxyethylene-based inhibitors bound to human immunodeficiency virus type 1 protease show that the inhibitors are present in two distinct orientations | | Descriptor: | HIV-1 PROTEASE, SKF 108738 PEPTIDE INHIBITOR | | Authors: | Murthy, K, Winborne, E.L, Minnich, M.D, Culp, J.S, Debouck, C. | | Deposit date: | 1992-09-21 | | Release date: | 1994-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures at 2.2-A resolution of hydroxyethylene-based inhibitors bound to human immunodeficiency virus type 1 protease show that the inhibitors are present in two distinct orientations.
J.Biol.Chem., 267, 1992
|
|
1HEG
 
 | | The crystal structures at 2.2 angstroms resolution of hydroxyethylene-based inhibitors bound to human immunodeficiency virus type 1 protease show that the inhibitors are present in two distinct orientations | | Descriptor: | HIV-1 PROTEASE, methyl N-{(4S,5S)-5-[(L-alanyl-L-alanyl)amino]-4-hydroxy-6-phenylhexanoyl}-L-valyl-L-valinate | | Authors: | Murthy, K, Winborne, E.L, Minnich, M.D, Culp, J.S, Debouck, C. | | Deposit date: | 1992-09-21 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures at 2.2-A resolution of hydroxyethylene-based inhibitors bound to human immunodeficiency virus type 1 protease show that the inhibitors are present in two distinct orientations.
J.Biol.Chem., 267, 1992
|
|
1HMV
 
 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | Authors: | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | Deposit date: | 1994-12-15 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1LAY
 
 | | CRYSTAL STRUCTURE OF CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CYTOMEGALOVIRUS PROTEASE | | Authors: | Qiu, X, Culp, J.S, Dilella, A.G, Hellmig, B, Hoog, S.S, Jason, C.A, Smith, W.W, Abdel-Meguid, S.S. | | Deposit date: | 1996-07-16 | | Release date: | 1997-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique fold and active site in cytomegalovirus protease.
Nature, 383, 1996
|
|
2P4E
 
 | | Crystal Structure of PCSK9 | | Descriptor: | MERCURY (II) ION, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Cunningham, D, Danley, D.E, Geoghegan, F.K, Griffor, M.C, Hawkins, J.L, Qiu, X. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1SIV
 
 | |
4EWS
 
 | |
2OBD
 
 | | Crystal Structure of Cholesteryl Ester Transfer Protein | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Qiu, X. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6MP6
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
6MPB
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
1HOS
 
 | |
5WBP
 
 | |
5WBM
 
 | |
5WBR
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-[(3S)-3-(hydroxymethyl)piperidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, GLYCEROL, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5WBO
 
 | |
5WBQ
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 2-ethyl-7-[(3S)-3-hydroxy-3-methylpyrrolidin-1-yl]-5-(trifluoromethyl)-1H-pyrrolo[3,2-b]pyridine-6-carbonitrile, CHLORIDE ION, Ketohexokinase, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5WBZ
 
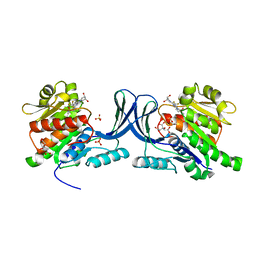 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 6-[(3S,4S)-3,4-dihydroxypyrrolidin-1-yl]-2-[(3S)-3-hydroxy-3-methylpyrrolidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, Ketohexokinase, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
