6OHI
 
 | | Crystal Structure of the Debrominase Bmp8 (Apo) | | Descriptor: | Debrominase Bmp8, SULFATE ION | | Authors: | Chekan, J.R, Moore, B.S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Bacterial Tetrabromopyrrole Debrominase Shares a Reductive Dehalogenation Strategy with Human Thyroid Deiodinase.
Biochemistry, 58, 2019
|
|
6OHJ
 
 | | Crystal Structure of the Debrominase Bmp8 C82A in Complex with 2,3,4-tribromopyrrole | | Descriptor: | 2,3,4-tribromo-1H-pyrrole, Debrominase Bmp8, SULFATE ION | | Authors: | Chekan, J.R, Moore, B.S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Bacterial Tetrabromopyrrole Debrominase Shares a Reductive Dehalogenation Strategy with Human Thyroid Deiodinase.
Biochemistry, 58, 2019
|
|
6VL0
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with GSPP and Mn2+ | | Descriptor: | DabA, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VKZ
 
 | |
6VL1
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with NGG and Mg2+ | | Descriptor: | DabA, MAGNESIUM ION, N-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-L-glutamic acid | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5BKE
 
 | | Crystal structure of AAD-2 in complex with Mn(II) and N-oxalylglycine | | Descriptor: | Alpha-ketoglutarate-dependent 2,4-dichlorophenoxyacetate dioxygenase, MANGANESE (II) ION, N-OXALYLGLYCINE, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2019-06-02 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5BKD
 
 | | Crystal structure of AAD-1 in complex with (R)-cyhalofop, Mn(II), and 2-oxoglutarate | | Descriptor: | (2R)-2-[4-(4-cyano-2-fluorophenoxy)phenoxy]propanoic acid, (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2019-06-02 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5BKC
 
 | | Crystal structure of AAD-1 in complex with (R)-diclofop, Mn(II), and 2-oxoglutarate | | Descriptor: | (2R)-2-{4-[(3,5-dichloropyridin-2-yl)oxy]phenoxy}propanoic acid, (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2019-06-02 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5BKB
 
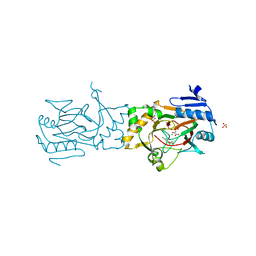 | | Crystal structure of AAD-1 in complex with (R)-dichlorprop, Mn(II), and 2-oxoglutarate | | Descriptor: | (2R)-2-(2,4-dichlorophenoxy)propanoic acid, (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2019-06-02 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5TXE
 
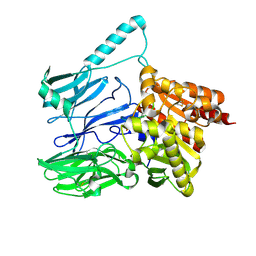 | |
5TXC
 
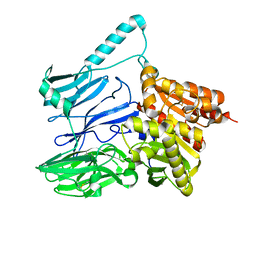 | | AtxE2 Isopeptidase - APO | | Descriptor: | AtxE2 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of the Lasso Peptide Isopeptidase Identifies a Topology for Processing Threaded Substrates.
J. Am. Chem. Soc., 138, 2016
|
|
5UW6
 
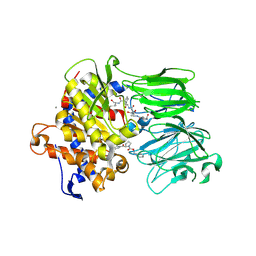 | | PCY1 in Complex with Follower Peptide and Covalent Inhibitor ZPP | | Descriptor: | CALCIUM ION, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Peptide cyclase 1, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V1U
 
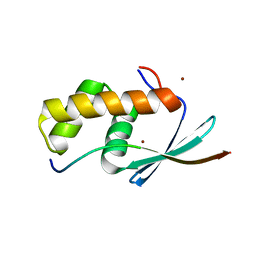 | | TbiB1 in Complex with the TbiA(beta) Leader Peptide | | Descriptor: | TbiA(beta) Thr(-5)Glu Leader, TbiB1, ZINC ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-03-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Steric complementarity directs sequence promiscuous leader binding in RiPP biosynthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5UW5
 
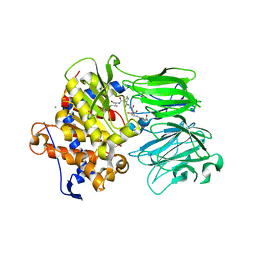 | | PCY1 H695A Variant in Complex with Follower Peptide | | Descriptor: | CACODYLATE ION, CALCIUM ION, Peptide cyclase 1, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UW7
 
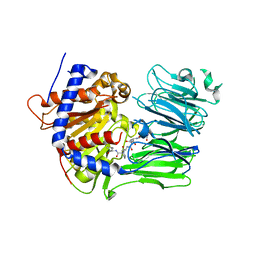 | | PCY1 Y481F Variant in Complex with Follower Peptide | | Descriptor: | MAGNESIUM ION, Peptide cyclase 1, Presegetalin A1 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UW3
 
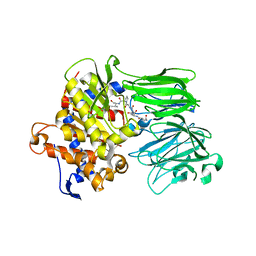 | | PCY1 in Complex with Follower Peptide | | Descriptor: | CACODYLATE ION, Peptide cyclase 1, Presegetalin A1 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4R9F
 
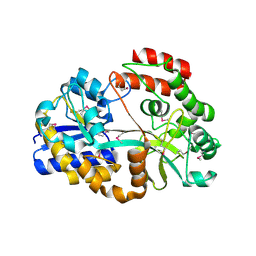 | | CpMnBP1 with Mannobiose Bound | | Descriptor: | MBP1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Chekan, J.R, Agarwal, V, Nair, S.K. | | Deposit date: | 2014-09-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Basis for Mannan Utilization by Caldanaerobius polysaccharolyticus Strain ATCC BAA-17.
J.Biol.Chem., 289, 2014
|
|
4R9G
 
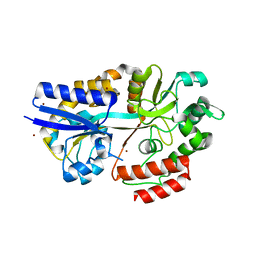 | | CpMnBP1 with Mannotriose Bound | | Descriptor: | MBP1, ZINC ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Chekan, J.R, Agarwal, V, Nair, S.K. | | Deposit date: | 2014-09-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Biochemical Basis for Mannan Utilization by Caldanaerobius polysaccharolyticus Strain ATCC BAA-17.
J.Biol.Chem., 289, 2014
|
|
5V1V
 
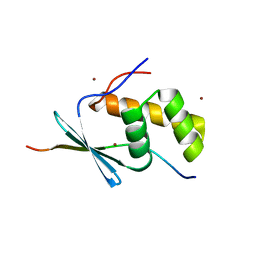 | | TbiB1 in Complex with the TbiA(alpha) Leader Peptide | | Descriptor: | TbiA(alpha) Leader Peptide, TbiB1, ZINC ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-03-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Steric complementarity directs sequence promiscuous leader binding in RiPP biosynthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5UZW
 
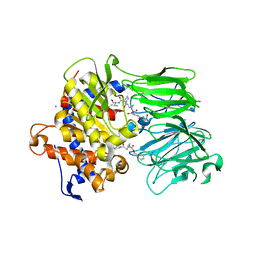 | |
4YAR
 
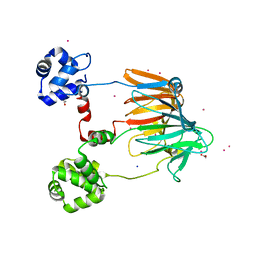 | | 2-Hydroxyethylphosphonate dioxygenase (HEPD) E176H | | Descriptor: | 2-hydroxyethylphosphonate dioxygenase, ACETATE ION, CADMIUM ION, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Common Late-Stage Intermediate in Catalysis by 2-Hydroxyethyl-phosphonate Dioxygenase and Methylphosphonate Synthase.
J.Am.Chem.Soc., 137, 2015
|
|
7M0O
 
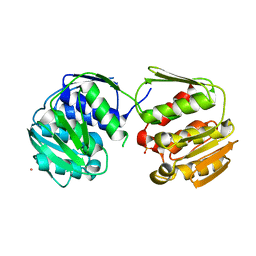 | | DGT-28 EPSPS | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, PHOSPHATE ION, POTASSIUM ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Characterization of a Glyphosate-Tolerant Enzyme from Streptomyces svecius : A Distinct Class of 5-Enolpyruvylshikimate-3-phosphate Synthases.
J.Agric.Food Chem., 69, 2021
|
|
5BK9
 
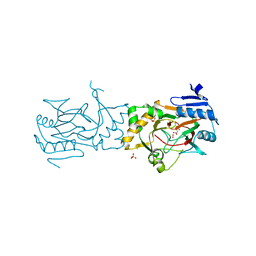 | | AAD-1 Bound to the Vanadyl Ion and Succinate | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, PHOSPHATE ION, SUCCINIC ACID, ... | | Authors: | Ongpipattanakul, C, Chekan, J.R. | | Deposit date: | 2019-06-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4MGS
 
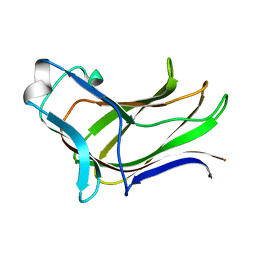 | | BiXyn10A CBM1 APO | | Descriptor: | Putative glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MGQ
 
 | | PbXyn10C CBM APO | | Descriptor: | CALCIUM ION, Glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
