1SXJ
 
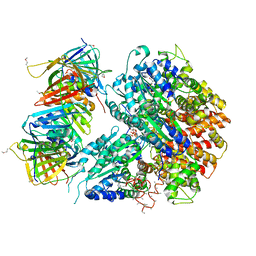 | | Crystal Structure of the Eukaryotic Clamp Loader (Replication Factor C, RFC) Bound to the DNA Sliding Clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Activator 1 37 kDa subunit, Activator 1 40 kDa subunit, ... | | Authors: | Bowman, G.D, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex.
Nature, 429, 2004
|
|
1F7S
 
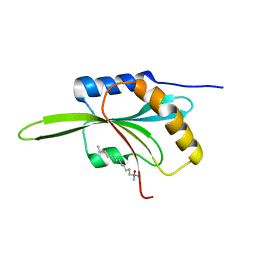 | | CRYSTAL STRUCTURE OF ADF1 FROM ARABIDOPSIS THALIANA | | Descriptor: | ACTIN DEPOLYMERIZING FACTOR (ADF), LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Bowman, G.D, Nodelman, I.M, Lindberg, U, Chua, N.H, Schutt, C.E. | | Deposit date: | 2000-06-27 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A comparative structural analysis of the ADF/cofilin family.
Proteins, 41, 2000
|
|
1G1J
 
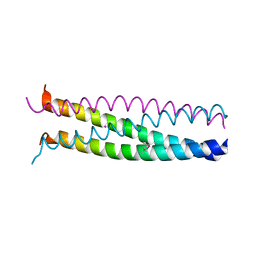 | |
1G1I
 
 | |
5J70
 
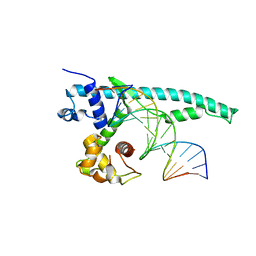 | | The Chd1 DNA-binding domain in complex with 17mer DNA duplex | | Descriptor: | Chromo domain-containing protein 1, DNA (5'-D(*CP*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*CP*G)-3') | | Authors: | Bowman, G.D, Jenkins, K.R, Hauk, G. | | Deposit date: | 2016-04-05 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Chd1 domains communicate across the DNA gyres of the nucleosome
To Be Published
|
|
4Y6C
 
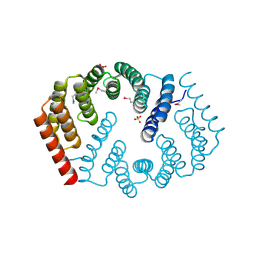 | | Q17M crystal structure of Podosopora anserina putative kinesin light chain nearly identical TPR-like repeats | | Descriptor: | ANSERINA PUTATIVE KINESIN LIGHT chain, SULFATE ION | | Authors: | Marold, J.D, Kavran, J.M, Bowman, G.D, Barrick, D. | | Deposit date: | 2015-02-12 | | Release date: | 2015-10-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | A Naturally Occurring Repeat Protein with High Internal Sequence Identity Defines a New Class of TPR-like Proteins.
Structure, 23, 2015
|
|
4Y6W
 
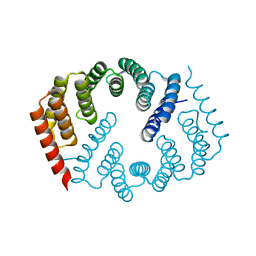 | |
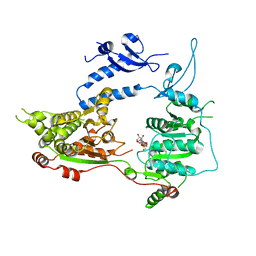
3MWY
 
 | |
3TED
 
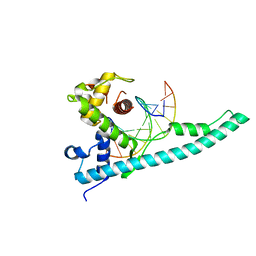 | | Crystal structure of the Chd1 DNA-binding domain in complex with a DNA duplex | | Descriptor: | 5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*AP*TP*AP*TP*AP*TP*GP*G)-3', Chromo domain-containing protein 1 | | Authors: | Sharma, A, Jenkins, K.R, Heroux, A, Bowman, G.D. | | Deposit date: | 2011-08-12 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-binding domain of Chd1 in complex with a DNA duplex
J.Biol.Chem., 2011
|
|
7SWY
 
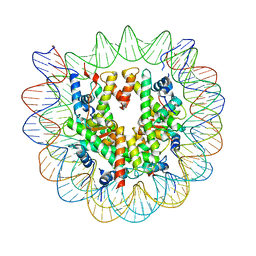 | | 2.6 A structure of a 40-601[TA-rich+1]-40 nucleosome | | Descriptor: | DNA Guide Strand, DNA Tracking Strand, Histone H2A, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TN2
 
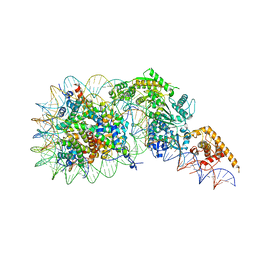 | | Composite model of a Chd1-nucleosome complex in the nucleotide-free state derived from 2.3A and 2.7A Cryo-EM maps | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | Deposit date: | 2022-01-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1D1J
 
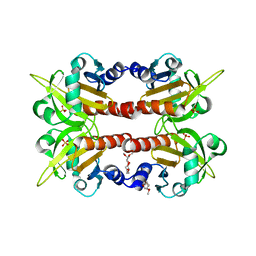 | | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, PROFILIN II, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Lindberg, U, Schutt, C.E. | | Deposit date: | 1999-09-17 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure determination of human profilin II: A comparative structural analysis of human profilins.
J.Mol.Biol., 294, 1999
|
|
3K4X
 
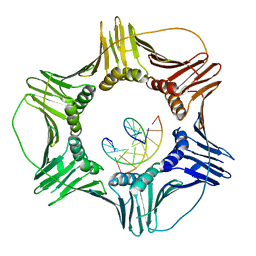 | | Eukaryotic Sliding Clamp PCNA Bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), Proliferating cell nuclear antigen | | Authors: | McNally, R, Kuriyan, J. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Analysis of the role of PCNA-DNA contacts during clamp loading.
Bmc Struct.Biol., 10, 2010
|
|
