5JUV
 
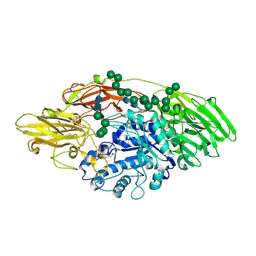 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-b-Galactopyranosyl galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-05-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
1LUF
 
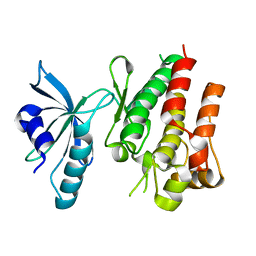 | | Crystal Structure of the MuSK Tyrosine Kinase: Insights into Receptor Autoregulation | | Descriptor: | muscle-specific tyrosine kinase receptor musk | | Authors: | Till, J.H, Becerra, M, Watty, A, Lu, Y, Ma, Y, Neubert, T.A, Burden, S.J, Hubbard, S.R. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation.
Structure, 10, 2002
|
|
5MGC
 
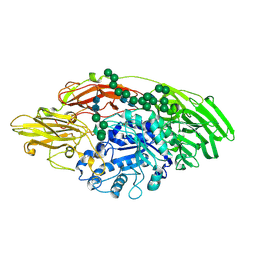 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 4-Galactosyl-lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Probable beta-galactosidase A, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
5MGD
 
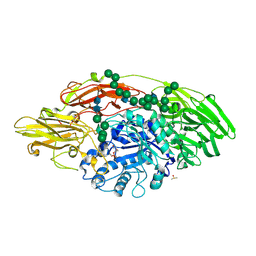 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-Galactosyl-lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
5IHR
 
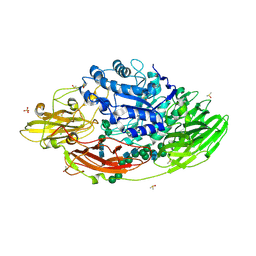 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH ALLOLACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-02-29 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
5IFP
 
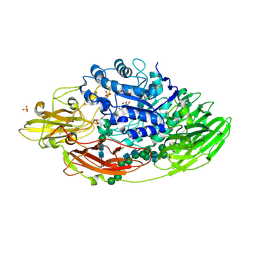 | | STRUCTURE OF BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-02-26 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
5IFT
 
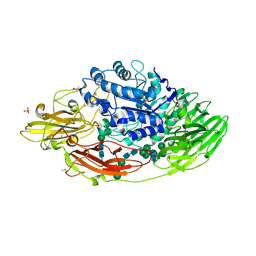 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 3-b-Galactopyranosyl glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-02-26 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
3OBA
 
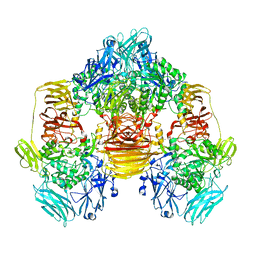 | | Structure of the beta-galactosidase from Kluyveromyces lactis | | Descriptor: | Beta-galactosidase, GLYCEROL, MANGANESE (III) ION | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Becerra, M, Gonzalez-Siso, I, Cerdan, M.E, Sanz-Aparicio, J. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase.
J.Struct.Biol., 177, 2012
|
|
3OB8
 
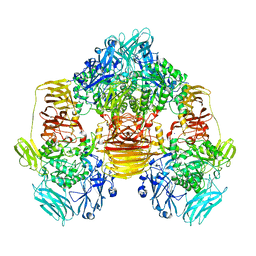 | | Structure of the beta-galactosidase from Kluyveromyces lactis in complex with galactose | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Becerra, M, Gonzalez-Siso, I, Cerdan, M.E, Sanz-Aparicio, J. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase.
J.Struct.Biol., 177, 2012
|
|
3LRK
 
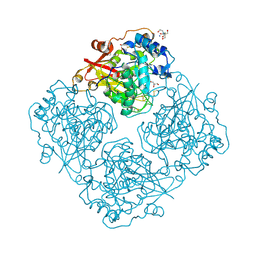 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
3LRM
 
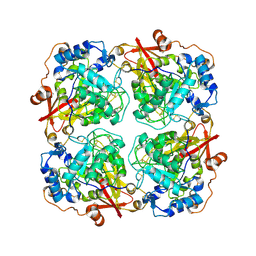 | | Structure of alfa-galactosidase from Saccharomyces cerevisiae with raffinose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
3LRL
 
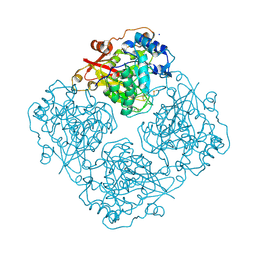 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae with melibiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
7NB5
 
 | |
7ATF
 
 | | Structure of EstD11 in complex with p-Nitrophenol | | Descriptor: | ACETATE ION, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AV5
 
 | | Structure of EstD11 in complex with Fluorescein | | Descriptor: | ACETATE ION, EstD11, FLUORESCIN, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AT0
 
 | |
7ATQ
 
 | | Structure of EstD11 in complex with cyclohexane carboxylic acid | | Descriptor: | ACETATE ION, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AT4
 
 | | Structure of EstD11 in complex with Naproxen | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, EstD11, FORMIC ACID | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AUY
 
 | |
7AT2
 
 | |
7AT3
 
 | | Structure of EstD11 in complex with Naproxen and methanol | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7ATD
 
 | | Structure of inactive EstD11 S144A in complex with methyl-naproxen | | Descriptor: | ACETATE ION, EstD11 S144A, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-29 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
