5F3K
 
 | | X-Ray Crystallographic Structure of hTrap1 N-terminal Domain-apo | | Descriptor: | Heat shock protein 75 kDa, mitochondrial | | Authors: | Sung, N, Lee, J, Kim, J, Chang, C, Joachimiak, A, Lee, S, Tsai, F.T.F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HPH
 
 | | Structure of TRAP1 fragment | | Descriptor: | GLYCEROL, Heat shock protein 75 kDa, mitochondrial, ... | | Authors: | Sung, N, Chang, C, Lee, S, Tsai, F.T.F. | | Deposit date: | 2016-01-20 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | 2.4 angstrom resolution crystal structure of human TRAP1NM, the Hsp90 paralog in the mitochondrial matrix.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5F5R
 
 | | TRAP1N-ADPNP | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, MAGNESIUM ION, ... | | Authors: | Tsai, F.T.F, Lee, S, Sung, N, Lee, J, Chang, C, Joachimiak, A. | | Deposit date: | 2015-12-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MHE
 
 | | Galphai3 co-crystallized with KB752 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(k) subunit alpha, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MHF
 
 | | Galphai3 co-crystallized with GIV/Girdin | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Girdin, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4HIN
 
 | | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L) | | Descriptor: | COPPER (II) ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L)
To be Published
|
|
2I89
 
 | | Structure of septuple mutant of Rat Outer Mitochondrial Membrane Cytochrome B5 | | Descriptor: | Cytochrome b5 type B, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Terzyan, S, Zhang, X.C, Benson, D.R, Wang, L, Sun, N. | | Deposit date: | 2006-09-01 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A histidine/tryptophan pi-stacking interaction stabilizes the heme-independent folding core of microsomal apocytochrome b5 relative to that of mitochondrial apocytochrome b5.
Biochemistry, 45, 2006
|
|
4HIL
 
 | | 1.25A Resolution Structure of Rat Type B Cytochrome b5 | | Descriptor: | Cytochrome b5 type B, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25A Resolution Structure of Rat Type B Cytochrome b5
To be Published
|
|
8H2K
 
 | |
8H2V
 
 | |
8H6H
 
 | |
8H2W
 
 | |
8HO9
 
 | |
8HO7
 
 | |
8HNU
 
 | |
8HO8
 
 | |
6EJP
 
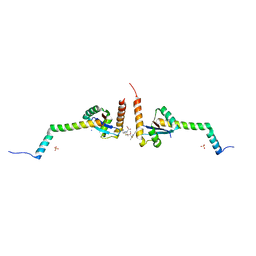 | | Yersinia YscU C-terminal fragment in complex with a synthetic compound | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Karlberg, T, Thorsell, A.G, Ho, O, Sunduru, N, Elofsson, M, Wolf-Watz, M, Schuler, H. | | Deposit date: | 2017-09-22 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Yersinia YscU C-terminal fragment in complex with a synthetic compound
To Be Published
|
|
4FD2
 
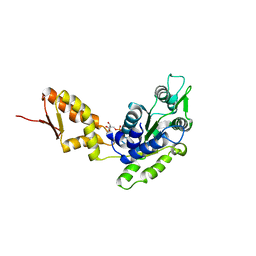 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FCV
 
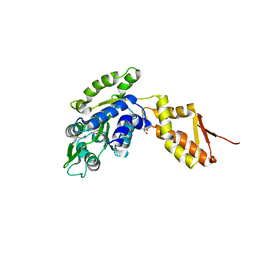 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FCT
 
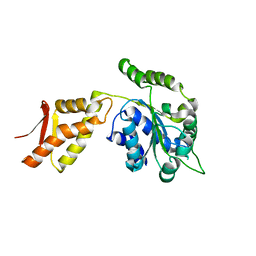 | |
4FCW
 
 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5V2W
 
 | | Crystal structure of a LuxS from salmonella typhi | | Descriptor: | S-ribosylhomocysteine lyase, ZINC ION | | Authors: | Perumal, P, Raina, R, Manoj Kumar, P, Arockisamy, A, SundaraBaalaji, N. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a LuxS from salmonella typhi
To Be Published
|
|
8WWT
 
 | |
8WUP
 
 | |
8WW5
 
 | |
