5HBW
 
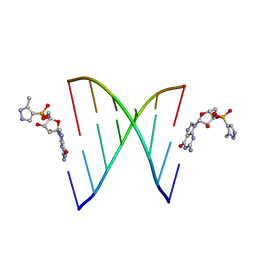 | | RNA primer-template complex with 2-methylimidazole-activated monomer analogue | | Descriptor: | RNA (5'-R(*(LCC)P*(TLN)P*(LCG)P*UP*AP*CP*A)-3'), [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-(3-methyl-1~{H}-pyrazol-4-yl)phosphinic acid | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2016-01-03 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
5HU4
 
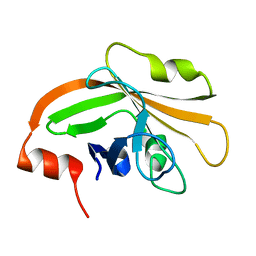 | | Cystal structure of listeria monocytogenes sortase A | | Descriptor: | Cysteine protease | | Authors: | Li, H. | | Deposit date: | 2016-01-27 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition of sortase A by chalcone prevents Listeria monocytogenes infection.
Biochem. Pharmacol., 106, 2016
|
|
7V9E
 
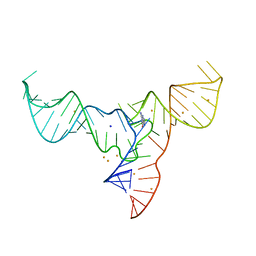 | | Crystal structure of a methyl transferase ribozyme | | Descriptor: | BARIUM ION, GUANINE, RNA (68-MER), ... | | Authors: | Deng, J, Lilley, D.M.J, Huang, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a methyltransferase ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
5FJ0
 
 | |
5FK2
 
 | |
5HZE
 
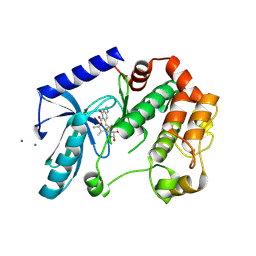 | | Mek1 adopts DFG-out conformation when bound to an analog of E6201. | | Descriptor: | (3S,4R,8S,9S,11E)-8,9,16-trihydroxy-3,4-dimethyl-14-(methylamino)-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION | | Authors: | Larsen, N.A, Bloudoff, K. | | Deposit date: | 2016-02-02 | | Release date: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mek1 adopts DFG-out conformation when bound to an analog of E6201.
To Be Published
|
|
5ZYQ
 
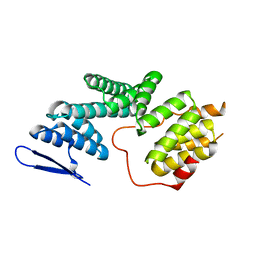 | | The Structure of Human PAF1/CTR9 complex | | Descriptor: | RNA polymerase-associated protein CTR9 homolog,RNA polymerase II-associated factor 1 homolog | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
7CLV
 
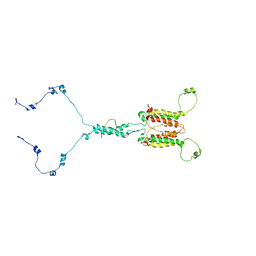 | | Solution structure of mitochondrial Tim23 channel in complex with a signaling peptide | | Descriptor: | COX4 isoform 1, TIM23 isoform 1 | | Authors: | Zhou, S, Ruan, M.S, Li, Y.Y, Yang, J, Richter, C, Schwalbe, H, Shen, B, Wang, J.F. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the voltage-gated Tim23 channel in complex with a mitochondrial presequence peptide.
Cell Res., 31, 2021
|
|
3S7V
 
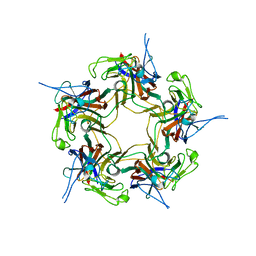 | | Unassembled KI Polyomavirus VP1 Pentamer | | Descriptor: | Major capsid protein VP1 | | Authors: | Neu, U, Stehle, T. | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of the major capsid proteins of the human KI and WU polyomaviruses.
J.Virol., 85, 2011
|
|
6LBK
 
 | |
5X41
 
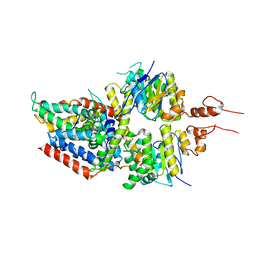 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
3TVM
 
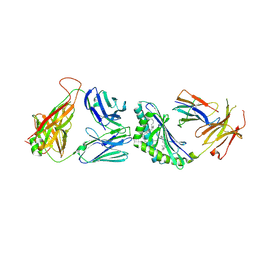 | | Structure of the mouse CD1d-SMC124-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Girardi, E, Li, Y, Zajonc, D.M. | | Deposit date: | 2011-09-20 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Glycolipids that Elicit IFN-gama-Biased Responses from Natural Killer T Cells
Chem.Biol., 18, 2011
|
|
6A0N
 
 | | The crystal structure of apo-Lpg2622 | | Descriptor: | Lpg2622 | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
2LCC
 
 | | Solution structure of RBBP1 chromobarrel domain | | Descriptor: | AT-rich interactive domain-containing protein 4A | | Authors: | Gong, W, Feng, Y. | | Deposit date: | 2011-04-28 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into recognition of methylated histone tails by retinoblastoma-binding protein 1.
J.Biol.Chem., 2012
|
|
6LYP
 
 | | Cryo-EM structure of AtMSL1 wild type | | Descriptor: | Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Sun, L. | | Deposit date: | 2020-02-15 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Insights into a Plant Mechanosensitive Ion Channel MSL1.
Cell Rep, 30, 2020
|
|
6AGS
 
 | |
4IRJ
 
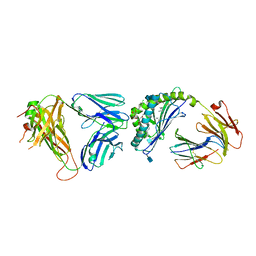 | | Structure of the mouse CD1d-4ClPhC-alpha-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Nemcovic, M, Zajonc, D.M. | | Deposit date: | 2013-01-14 | | Release date: | 2013-08-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enhanced TCR footprint by a novel glycolipid increases NKT-dependent tumor protection.
J.Immunol., 191, 2013
|
|
7DYR
 
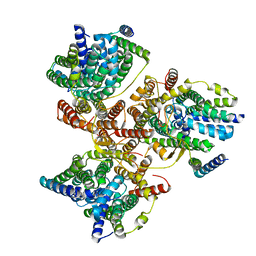 | |
6B0B
 
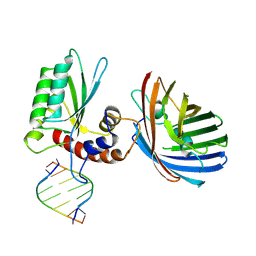 | | Crystal structure of human APOBEC3H | | Descriptor: | APOBEC3H, MCherry, RNA (5'-R(*UP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Shaban, N.M, Shi, K, Banerjee, S, Harris, R.S, Aihara, H. | | Deposit date: | 2017-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.2800622 Å) | | Cite: | The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism.
Mol. Cell, 69, 2018
|
|
5Y9V
 
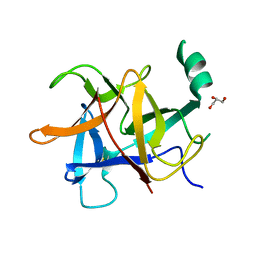 | | Crystal structure of diamondback moth ryanodine receptor N-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, Ryanodine receptor 1 | | Authors: | Lin, L, Yuchi, Z. | | Deposit date: | 2017-08-28 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Crystal structure of ryanodine receptor N-terminal domain from Plutella xylostella reveals two potential species-specific insecticide-targeting sites.
Insect Biochem. Mol. Biol., 92, 2017
|
|
5YOJ
 
 | | Structure of A17 HIV-1 Protease in Complex with Inhibitor KNI-1657 | | Descriptor: | (4R)-N-[(2,6-dimethylphenyl)methyl]-3-[(2S,3S)-3-[[(2S)-2-[(7-methoxy-1-benzofuran-2-yl)carbonylamino]-2-[(3R)-oxolan-3 -yl]ethanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, A17 HIV-1 protease, GLYCEROL | | Authors: | Adachi, M, Hidaka, K, Kuroki, R, Kiso, Y. | | Deposit date: | 2017-10-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Highly Potent Human Immunodeficiency Virus Type-1 Protease Inhibitors against Lopinavir and Darunavir Resistant Viruses from Allophenylnorstatine-Based Peptidomimetics with P2 Tetrahydrofuranylglycine.
J. Med. Chem., 61, 2018
|
|
6MJH
 
 | |
6BBO
 
 | | Crystal structure of human APOBEC3H/RNA complex | | Descriptor: | APOBEC3H, GLYCEROL, MCherry fluorescent protein, ... | | Authors: | Shaban, N.M, Shi, K, Banerjee, S, Harris, R.S, Aihara, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.428 Å) | | Cite: | The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism.
Mol. Cell, 69, 2018
|
|
5YR3
 
 | | Structure of sfYFP66BPA | | Descriptor: | Yellow fluorescent protein | | Authors: | Kang, F, Wang, L, Wang, J. | | Deposit date: | 2017-11-08 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | sfYFP66BPA
To Be Published
|
|
3TA3
 
 | | Structure of the mouse CD1d-Glc-DAG-s2-iNKT TCR complex | | Descriptor: | (2S)-1-(alpha-D-glucopyranosyloxy)-3-(hexadecanoyloxy)propan-2-yl (11Z)-octadec-11-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unique Interplay between Sugar and Lipid in Determining the Antigenic Potency of Bacterial Antigens for NKT Cells.
Plos Biol., 9, 2011
|
|
