7WYB
 
 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
7WY8
 
 | | ADGRL3/Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling
Mol.Cell, 82, 2022
|
|
7X10
 
 | | ADGRL3/miniG12 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7OFK
 
 | | Ligand complex of RORg LBD | | Descriptor: | (1~{R})-2-ethanoyl-~{N}-[4-[1,1,1,3,3,3-hexakis(fluoranyl)-2-oxidanyl-propan-2-yl]phenyl]-5-methylsulfonyl-1,3-dihydroisoindole-1-carboxamide, DIMETHYL SULFOXIDE, Nuclear receptor ROR-gamma, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | AZD0284, a Potent, Selective, and Orally Bioavailable Inverse Agonist of Retinoic Acid Receptor-Related Orphan Receptor C2.
J.Med.Chem., 64, 2021
|
|
7OFI
 
 | | Ligand complex of RORg LBD | | Descriptor: | (2~{R})-2-(4-ethylsulfonylphenyl)-~{N}-[4-[1,1,1,3,3,3-hexakis(fluoranyl)-2-oxidanyl-propan-2-yl]phenyl]-~{N}'-methyl-butanediamide, DIMETHYL SULFOXIDE, Nuclear receptor ROR-gamma, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | AZD0284, a Potent, Selective, and Orally Bioavailable Inverse Agonist of Retinoic Acid Receptor-Related Orphan Receptor C2.
J.Med.Chem., 64, 2021
|
|
7N61
 
 | | structure of C2 projections and MIPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAP147, FAP178, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7N6G
 
 | | C1 of central pair | | Descriptor: | CPC1, Calmodulin, DPY30, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6LOL
 
 | | The crystal structure of full length IpaH9.8 | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Ye, Y, Huang, H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
8Q7E
 
 | |
8Q7H
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8RHZ
 
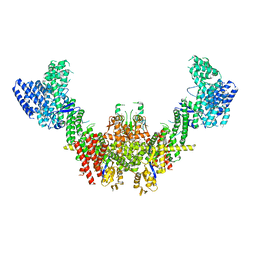 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2023-12-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5APJ
 
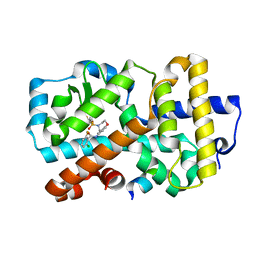 | | Ligand complex of RORg LBD | | Descriptor: | 2-CHLORO-6-FLUORO-N-[4-[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL-3,5-DIHYDRO-2H-1,4-BENZOXAZEPIN-7-YL]BENZAMIDE, NUCLEAR RECEPTOR COACTIVATOR 2, NUCLEAR RECEPTOR ROR-GAMMA, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
5APK
 
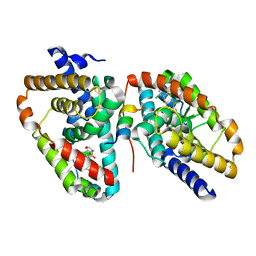 | | Ligand complex of RORg LBD | | Descriptor: | 2-CHLORO-6-FLUORO-N-[4-[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL-3,5-DIHYDRO-2H-1,4-BENZOXAZEPIN-7-YL]BENZAMIDE, NUCLEAR RECEPTOR ROR-GAMMA | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
4FFW
 
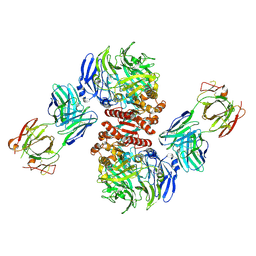 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with Fab + sitagliptin | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, Dipeptidyl peptidase 4, Fab heavy chain, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
1JL4
 
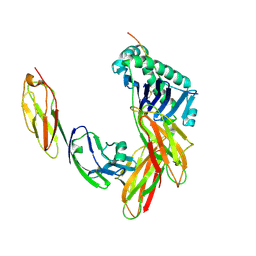 | | CRYSTAL STRUCTURE OF THE HUMAN CD4 N-TERMINAL TWO DOMAIN FRAGMENT COMPLEXED TO A CLASS II MHC MOLECULE | | Descriptor: | H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, A-K ALPHA CHAIN, A-K BETA CHAIN, ... | | Authors: | Wang, J.-H, Meijers, R, Reinherz, E.L. | | Deposit date: | 2001-07-15 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4FFV
 
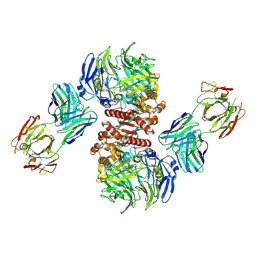 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with 11A19 Fab | | Descriptor: | 11A19 Fab heavy chain, 11A19 Fab light chain, Dipeptidyl peptidase 4 | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
7S5H
 
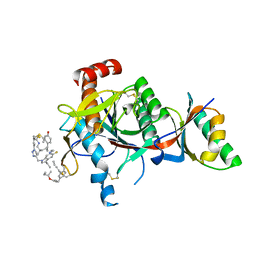 | | PCSK9(deltaCRD) in complex with cyclic peptide 35 | | Descriptor: | (2E)-but-2-ene-1,4-diol, Pro-peptide from Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Orth, P. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | A Series of Novel, Highly Potent, and Orally Bioavailable Next-Generation Tricyclic Peptide PCSK9 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7S5G
 
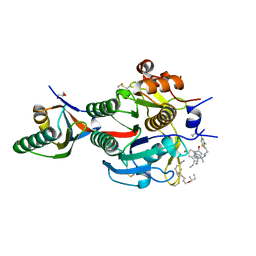 | | PCSK9 in complex with compound 19 | | Descriptor: | (2E)-but-2-ene-1,4-diol, GLYCEROL, Propeptide of Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Orth, P. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | A Series of Novel, Highly Potent, and Orally Bioavailable Next-Generation Tricyclic Peptide PCSK9 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6V40
 
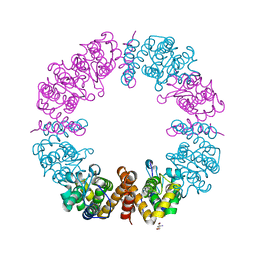 | | Structure of Salmonella Typhi TtsA | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PG_binding_3 domain-containing protein | | Authors: | Galan, J.E, Lara-Tejero, M. | | Deposit date: | 2019-11-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Mechanisms of substrate recognition by a typhoid toxin secretion-associated muramidase.
Elife, 9, 2020
|
|
7T7L
 
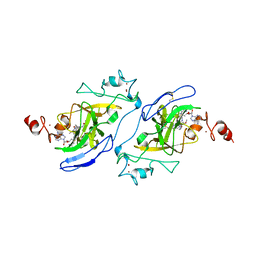 | | Structure of human G9a SET-domain (EHMT2) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
7T7M
 
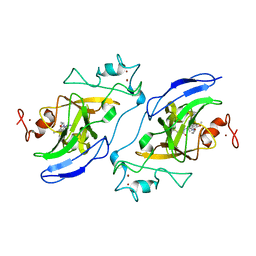 | | Structure of human GLP SET-domain (EHMT1) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
6WJ6
 
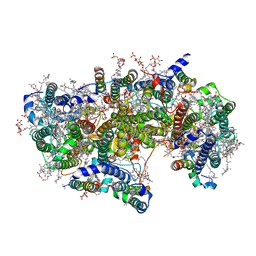 | | Cryo-EM structure of apo-Photosystem II from Synechocystis sp. PCC 6803 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gisriel, C.J. | | Deposit date: | 2020-04-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM Structure of Monomeric Photosystem II from Synechocystis sp. PCC 6803 Lacking the Water-Oxidation Complex
Joule, 2020
|
|
7YDM
 
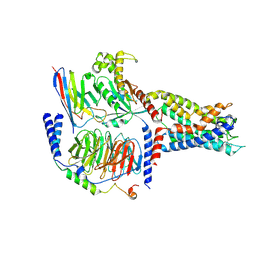 | | Cryo-EM structure of CD97/Gq complex | | Descriptor: | Adhesion G protein-coupled receptor E5 subunit beta, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis of CD97 activation and G-protein coupling.
Cell Chem Biol, 30, 2023
|
|
7YDP
 
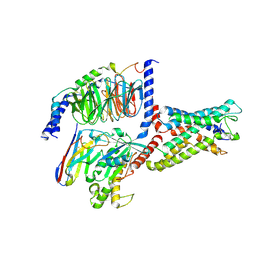 | | Cryo-EM structure of CD97/miniGs complex | | Descriptor: | Adhesion G protein-coupled receptor E5 subunit beta, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of CD97 activation and G-protein coupling.
Cell Chem Biol, 30, 2023
|
|
7YDH
 
 | | Cryo EM structure of CD97/miniG13 complex | | Descriptor: | Adhesion G protein-coupled receptor E5 subunit beta, G protein subunit 13 (Gi2-mini-G13 chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of CD97 activation and G-protein coupling.
Cell Chem Biol, 30, 2023
|
|
