6K63
 
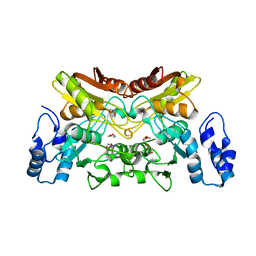 | | The crystal structure of cytidine deaminase from Klebsiella pneumoniae | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Cytidine deaminase, ZINC ION | | Authors: | Liu, W, Shang, F, Lan, J, Chen, Y, Wang, L, Xu, Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | Biochemical and structural analysis of the Klebsiella pneumoniae cytidine deaminase CDA.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
6K2Q
 
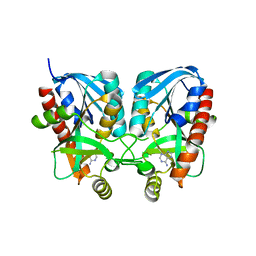 | | Aeromonas hydrophila MtaN-2 complexed with adenine | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE | | Authors: | Chen, J, Liu, W, Wang, L, Shang, F, Lan, J, Chen, Y, Xu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aeromonas hydrophila Cytoplasmic 5'-Methylthioadenosine/S-Adenosylhomocysteine Nucleosidase.
Biochemistry, 58, 2019
|
|
4R99
 
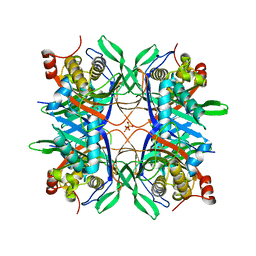 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | SULFATE ION, Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4R8X
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4YFF
 
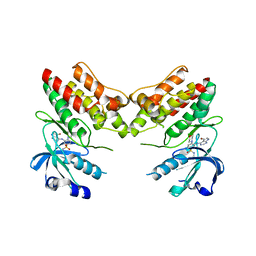 | | TNNI3K complexed with inhibitor 2 | | Descriptor: | 3-[(5-bromo-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-N-methyl-4-(morpholin-4-yl)benzenesulfonamide, Serine/threonine-protein kinase TNNI3K | | Authors: | Shewchuk, L.M, Wang, L, Lawhorn, B.G. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Identification of Purines and 7-Deazapurines as Potent and Selective Type I Inhibitors of Troponin I-Interacting Kinase (TNNI3K).
J.Med.Chem., 58, 2015
|
|
8I13
 
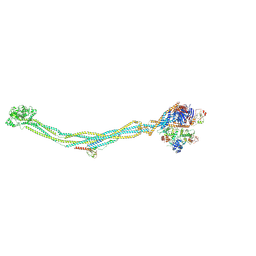 | | Cryo-EM structure of 6-subunit Smc5/6 | | Descriptor: | MMS21 isoform 1, NSE3 isoform 1, Non-structural maintenance of chromosomes element 1 homolog, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4U
 
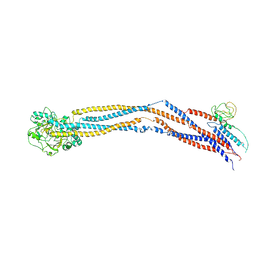 | | Cryo-EM structure of 5-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4V
 
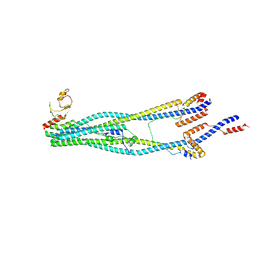 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4W
 
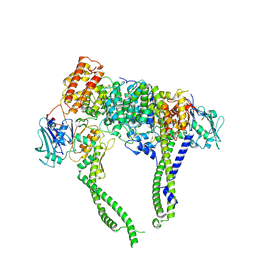 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4X
 
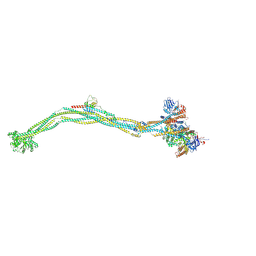 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4YFI
 
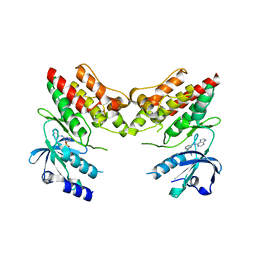 | | TNNI3K complexed with inhibitor 1 | | Descriptor: | N-methyl-3-(9H-purin-6-ylamino)benzenesulfonamide, Serine/threonine-protein kinase TNNI3K | | Authors: | Shewchuk, L.M, Wang, L, Lawhorn, B.G. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of Purines and 7-Deazapurines as Potent and Selective Type I Inhibitors of Troponin I-Interacting Kinase (TNNI3K).
J.Med.Chem., 58, 2015
|
|
4WUM
 
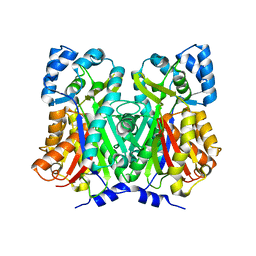 | |
5WXB
 
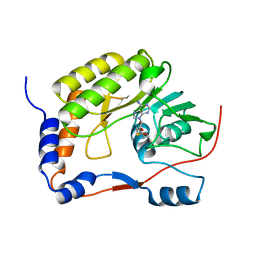 | |
3R7W
 
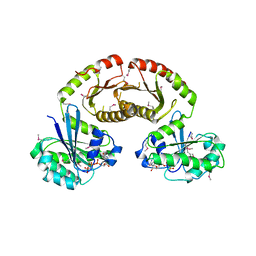 | | Crystal Structure of Gtr1p-Gtr2p complex | | Descriptor: | GTP-binding protein GTR1, GTP-binding protein GTR2, MAGNESIUM ION, ... | | Authors: | Gong, R, Li, L, Liu, Y, Wang, P, Yang, H, Wang, L, Cheng, J, Guan, K.L, Xu, Y. | | Deposit date: | 2011-03-23 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation
Genes Dev., 25, 2011
|
|
2ZGJ
 
 | | Crystal Structure of D86N-GzmM Complexed with Its Optimal Synthesized Substrate | | Descriptor: | Granzyme M, SSGKVPLS, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ZGH
 
 | | Crystal Structure of active granzyme M bound to its product | | Descriptor: | Granzyme M, SSGKVPL, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
5GJU
 
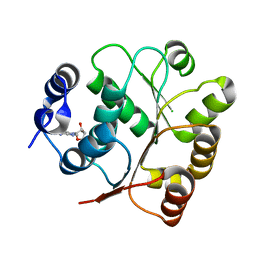 | | DEAD-box RNA helicase | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Li, F, Wang, L, Shi, Y. | | Deposit date: | 2016-07-02 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
5GI4
 
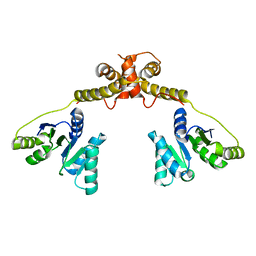 | | DEAD-box RNA helicase | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Wang, L, Li, F, Wu, L, Shi, Y. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
2ZGC
 
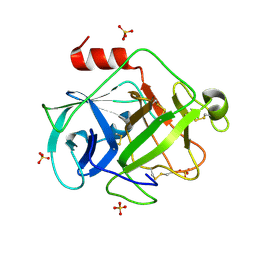 | | Crystal Structure of Active Human Granzyme M | | Descriptor: | Granzyme M, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ZKS
 
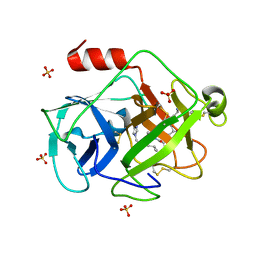 | | Structural insights into the proteolytic machinery of apoptosis-inducing Granzyme M | | Descriptor: | Granzyme M, SULFATE ION, hGzmM inhibitor | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Yang, X, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-03-28 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
4KFP
 
 | | Identification of 2,3-dihydro-1H-pyrrolo[3,4-c]pyridine-derived Ureas as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[1-(tetrahydro-2H-pyran-4-yl)piperidin-4-yl]sulfonyl}benzyl)-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Dragovich, P.S, Bair, K.W, Baumeister, T, Ho, Y, Liederer, B.M, Liu, X, O'Brien, T, Oeh, J, Sampath, D, Skelton, N, Wang, L, Wang, W, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X. | | Deposit date: | 2013-04-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Identification of 2,3-dihydro-1H-pyrrolo[3,4-c]pyridine-derived ureas as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8WJL
 
 | | Cryo-EM structure of 6-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.15 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
