5JJU
 
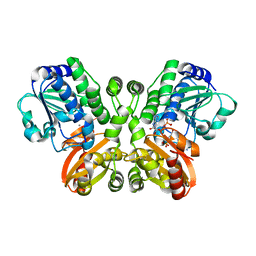 | | Crystal structure of Rv2837c complexed with 5'-pApA and 5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA (5'-R(P*AP*A)-3'), ... | | Authors: | Wang, F, He, Q, Liu, S, Gu, L. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
5GT2
 
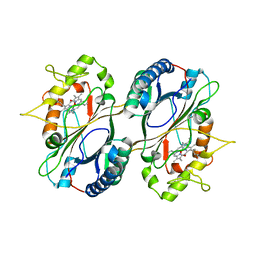 | | Crystal Structure and Biochemical Features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Probable deferrochelatase/peroxidase YfeX | | Authors: | Ma, Y.L, Yuan, Z.G, Liu, S, Wang, J.X, Gu, L.C, Liu, X.H. | | Deposit date: | 2016-08-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure and biochemical features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) play divergent roles toward different substrates
Biochem. Biophys. Res. Commun., 484, 2017
|
|
1MCT
 
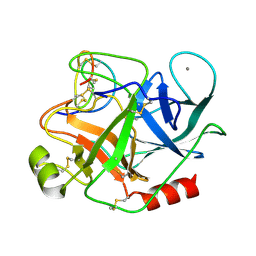 | | THE REFINED 1.6 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PORCINE BETA-TRYPSIN AND MCTI-A, A TRYPSIN INHIBITOR OF SQUASH FAMILY | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, TRYPSIN INHIBITOR A | | Authors: | Huang, Q, Liu, S, Tang, Y. | | Deposit date: | 1992-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined 1.6 A resolution crystal structure of the complex formed between porcine beta-trypsin and MCTI-A, a trypsin inhibitor of the squash family. Detailed comparison with bovine beta-trypsin and its complex.
J.Mol.Biol., 229, 1993
|
|
1MRH
 
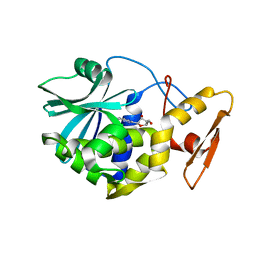 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRK
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
4ZMU
 
 | | Dcsbis, a diguanylate cyclase from Pseudomonas aeruginosa | | Descriptor: | diguanylate cyclase | | Authors: | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | crystal structure of Dcsbis from Pseudomonas aeruginosa
To Be Published
|
|
4ZMM
 
 | | GGDEF domain of Dcsbis complexed with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), diguanylate cyclase | | Authors: | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of Dcsbis GGDEF domain complexed with c-di-GMP
To Be Published
|
|
1MRJ
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
3QIC
 
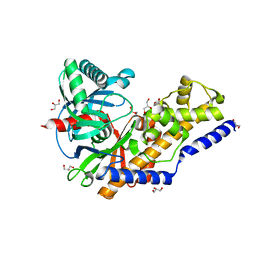 | | The structure of human glucokinase E339K mutation | | Descriptor: | GLYCEROL, Glucokinase, alpha-D-glucopyranose | | Authors: | Liu, Q, Liu, S, Liu, J. | | Deposit date: | 2011-01-27 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of E339K mutated human glucokinase reveals changes in the ATP binding site.
Febs Lett., 585, 2011
|
|
1MRI
 
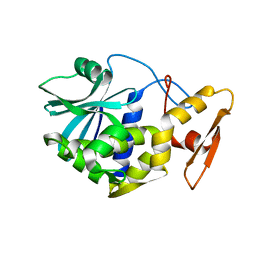 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRG
 
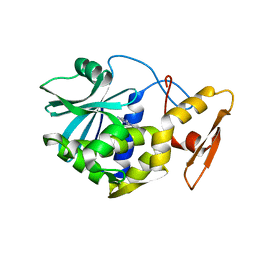 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
6PSN
 
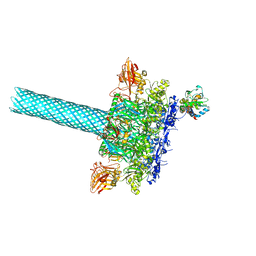 | | Anthrax toxin protective antigen channels bound to lethal factor | | Descriptor: | CALCIUM ION, Lethal factor, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
3TEF
 
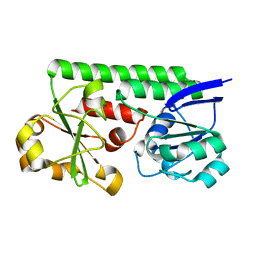 | | Crystal Structure of the Periplasmic Catecholate-Siderophore Binding Protein VctP from Vibrio Cholerae | | Descriptor: | Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | Authors: | Liu, X, Wang, Z, Liu, S, Li, N, Chen, Y, Zhu, C, Zhu, D, Wei, T, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-13 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of periplasmic catecholate-siderophore binding protein VctP from Vibrio cholerae at 1.7 A resolution
Febs Lett., 586, 2012
|
|
8E40
 
 | | Full-length APOBEC3G in complex with HIV-1 Vif, CBF-beta, and fork RNA | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, RNA, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Liu, S, Yang, H, Shiriaeva, A, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for HIV-1 antagonism of host APOBEC3G via Cullin E3 ligase.
Sci Adv, 9, 2023
|
|
1KBL
 
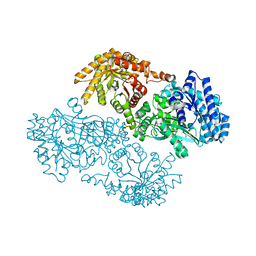 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | AMMONIUM ION, PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C, Liu, S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme-phosphonopyruvate complex, and mutant analysis
Biochemistry, 41, 2002
|
|
1EPT
 
 | | REFINED 1.8 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF PORCINE EPSILON-TRYPSIN | | Descriptor: | CALCIUM ION, PORCINE E-TRYPSIN | | Authors: | Huang, Q, Wang, Z, Li, Y, Liu, S, Tang, Y. | | Deposit date: | 1994-06-07 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A resolution crystal structure of the porcine epsilon-trypsin.
Biochim.Biophys.Acta, 1209, 1994
|
|
6WO0
 
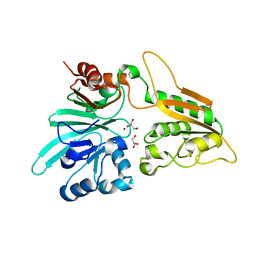 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | Descriptor: | GLYCEROL, Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
6WNL
 
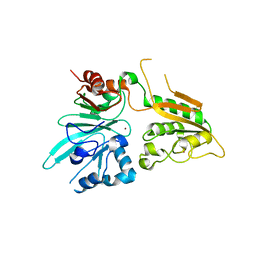 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | Descriptor: | Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
6WJJ
 
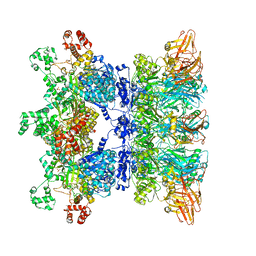 | | Anthrax octamer prechannel bound to full-length lethal factor | | Descriptor: | CALCIUM ION, Lethal factor, Protective antigen, ... | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
1SMF
 
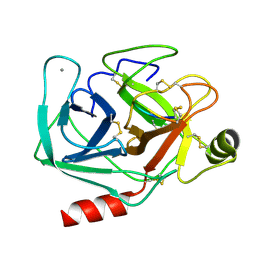 | | Studies on an artificial trypsin inhibitor peptide derived from the mung bean inhibitor | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, TRYPSIN | | Authors: | Huang, Q, Li, Y, Zhang, S, Liu, S, Tang, Y, Qi, C. | | Deposit date: | 1992-10-24 | | Release date: | 1994-07-31 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin.
J.Biochem.(Tokyo), 116, 1994
|
|
5HRA
 
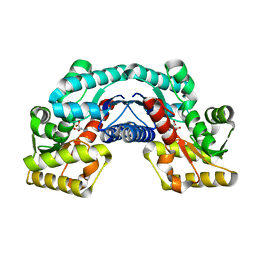 | | Crystal structure of an aspartate/glutamate racemase in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
5LQY
 
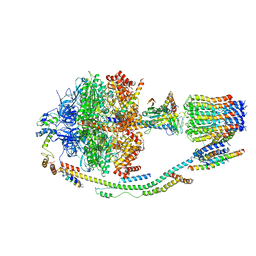 | | Structure of F-ATPase from Pichia angusta, in state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5HRC
 
 | | Crystal structure of an aspartate/glutamate racemase in complex with L-aspartate | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ASPARTIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
5LQX
 
 | | Structure of F-ATPase from Pichia angusta, state3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase AAP1 subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LQZ
 
 | | Structure of F-ATPase from Pichia angusta, state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
