5JRE
 
 | | Crystal structure of NeC3PO in complex with ssDNA. | | Descriptor: | 9-METHYL-9H-PURIN-6-AMINE, ADENINE, NEQ131, ... | | Authors: | Gan, J, Zhang, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
5JRC
 
 | |
9LCR
 
 | | Clostridium botulinum OLE RNA dimer | | Descriptor: | MAGNESIUM ION, RNA (578-MER) | | Authors: | Jia, X.Y, Wang, L, Su, Z.M. | | Deposit date: | 2025-01-05 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM reveals mechanisms of natural RNA multivalency.
Science, 388, 2025
|
|
5JR9
 
 | | Crystal structure of apo-NeC3PO | | Descriptor: | NEQ131 | | Authors: | Zhang, J, Gan, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
1J0Q
 
 | | Solution Structure of Oxidized Bovine Microsomal Cytochrome b5 mutant V61H | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, H, Huang, Z, Cao, C, Zhang, Q, Wang, Y.-H, Ma, J.-B, Xue, L.-L. | | Deposit date: | 2002-11-20 | | Release date: | 2003-08-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the oxidized bovine microsomal cytochrome b5 mutant V61H
Biochem.Biophys.Res.Commun., 307, 2003
|
|
7RUN
 
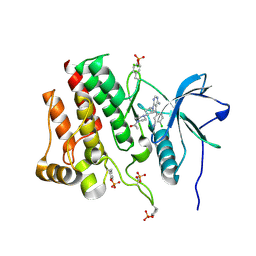 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with a pyrrolo[2,3-d]pyrimidine inhibitor. | | Descriptor: | 1-(4-{(1s,3s)-3-[4-amino-5-(3-amino-4-chlorophenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl]cyclobutyl}piperazin-1-yl)ethan-1-one, CHLORIDE ION, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2021-08-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Antitarget Selectivity and Tolerability of Novel Pyrrolo[2,3- d ]pyrimidine RET Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
9VD9
 
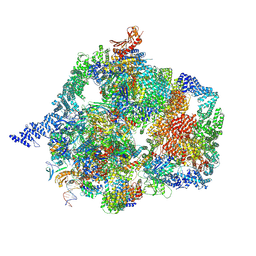 | |
5TP0
 
 | | Human mesotrypsin in complex with diminazene | | Descriptor: | BERENIL, CALCIUM ION, SULFATE ION, ... | | Authors: | Kayode, O, Soares, A, Radisky, E.S. | | Deposit date: | 2016-10-19 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule inhibitors of mesotrypsin from a structure-based docking screen.
PLoS ONE, 12, 2017
|
|
5XUA
 
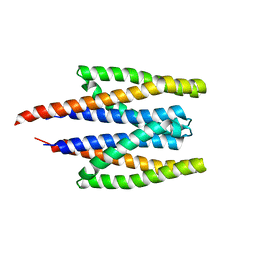 | |
5XMA
 
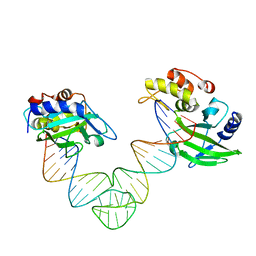 | |
5XUB
 
 | |
5XM9
 
 | |
5MZ3
 
 | | P38 ALPHA MUTANT C162S IN COMPLEX WITH CMPD2 [N-(4-Methyl-3-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)-3-(trifluoromethyl)benzamide] | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Cowan-Jacob, S.W, Scheufler, C. | | Deposit date: | 2017-01-30 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2FMX
 
 | | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+) | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Bian, C, Yuan, C, Li, Y, Huang, M. | | Deposit date: | 2006-01-10 | | Release date: | 2006-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+)
Biochem.Biophys.Res.Commun., 348, 2006
|
|
2FA9
 
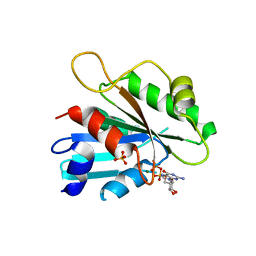 | | The crystal structure of Sar1[H79G]-GDP provides insight into the coat-controlled GTP hydrolysis in the disassembly of COP II | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Huang, M, Yuan, C, Bian, C, Hou, X. | | Deposit date: | 2005-12-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Sar1[H79G]-GDP Which Provides Insight into the Coat-controlled GTP Hydrolysis in the Disassembly of COP II
Chin.J.Struct.Chem., 25, 2006
|
|
6LQC
 
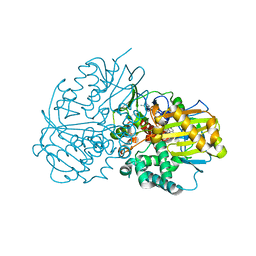 | | Crystal structure of Cyclohexylamine Oxidase from Erythrobacteraceae bacterium | | Descriptor: | Cyclohexylamine Oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Huang, Z.D. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Asymmetric Synthesis of a Key Dextromethorphan Intermediate and Its Analogues Enabled by a New Cyclohexylamine Oxidase: Enzyme Discovery, Reaction Development, and Mechanistic Insight.
J.Org.Chem., 85, 2020
|
|
6M3M
 
 | |
3B9L
 
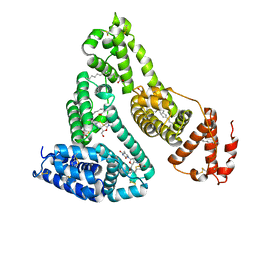 | | Human serum albumin complexed with myristate and AZT | | Descriptor: | 3'-azido-3'-deoxythymidine, MYRISTIC ACID, Serum albumin | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
3B9M
 
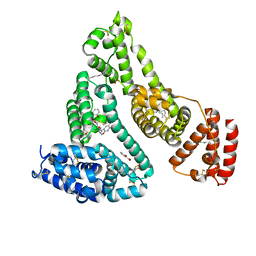 | | Human serum albumin complexed with myristate, 3'-azido-3'-deoxythymidine (AZT) and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, 3'-azido-3'-deoxythymidine, MYRISTIC ACID, ... | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
6LQL
 
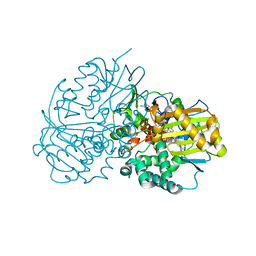 | | Complex structure of CHAO with product from Erythrobacteraceae bacterium | | Descriptor: | 1-[(4-methoxyphenyl)methyl]-3,4,5,6,7,8-hexahydroisoquinoline, Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Huang, Z.D. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asymmetric Synthesis of a Key Dextromethorphan Intermediate and Its Analogues Enabled by a New Cyclohexylamine Oxidase: Enzyme Discovery, Reaction Development, and Mechanistic Insight.
J.Org.Chem., 85, 2020
|
|
5UI0
 
 | |
8HLP
 
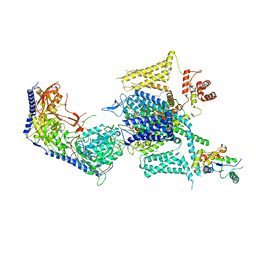 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2024-04-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMB
 
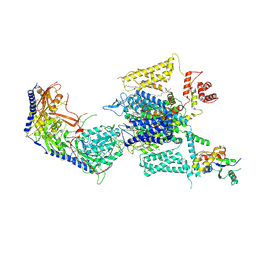 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with benidipine (BEN) | | Descriptor: | (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-4-(3-nitrophenyl)piperidine-3,5-dicarboxylate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMA
 
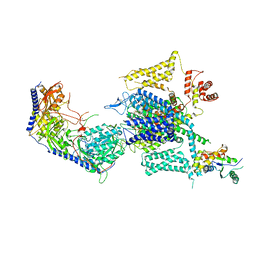 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
1I5U
 
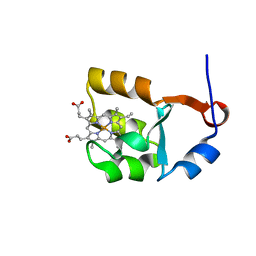 | | SOLUTION STRUCTURE OF CYTOCHROME B5 TRIPLE MUTANT (E48A/E56A/D60A) | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qian, C, Yao, Y, Tang, W, Wang, J, Zhongxian, H. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c.
Protein Sci., 10, 2001
|
|
