7CGG
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5A (5'-R(P*UP*GP*UP*AP*AP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGH
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5C (5'-R(P*UP*GP*UP*AP*CP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGK
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5A (5'-R(P*UP*GP*UP*AP*AP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGL
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5C (5'-R(P*UP*GP*UP*AP*CP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGI
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5U (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGF
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5G (5'-R(P*UP*GP*UP*AP*GP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGM
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5U (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGJ
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5G (5'-R(P*UP*GP*UP*AP*GP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.546 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7D54
 
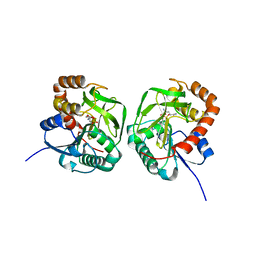 | | Crstal structure MsGATase with Gln | | Descriptor: | GLUTAMINE, Glutamine amidotransferase class-I | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D4R
 
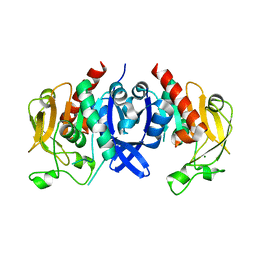 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D50
 
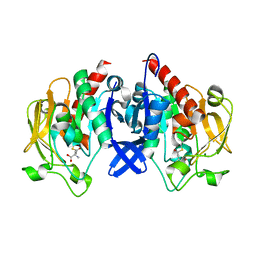 | | SpuA mutant - H221N with glutamyl-thioester | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D53
 
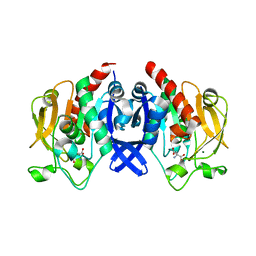 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8XN7
 
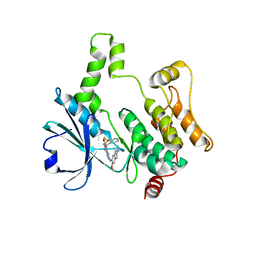 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 9f | | Descriptor: | 5-amino-2-((6-methoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-7-yl)amino)-8-(2-(trifluoromethyl)benzyl)pyrido[2,3-d]pyrimidin-7(8H)-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Huang, W.X, Liu, R, Ding, K. | | Deposit date: | 2023-12-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of 5-aminopyrido[2,3-d]pyrimidin-7(8H)-one derivatives as new hematopoietic progenitor kinase 1 (HPK1) inhibitors.
Eur.J.Med.Chem., 269, 2024
|
|
6M02
 
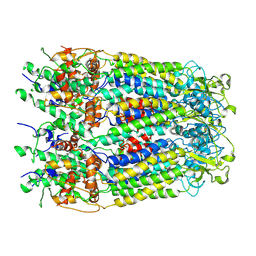 | | cryo-EM structure of human Pannexin 1 channel | | Descriptor: | Pannexin-1 | | Authors: | Ronggui, Q, Lili, D, Jilin, Z, Xuekui, Y, Lei, W, Shujia, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of human heptameric Pannexin 1 channel.
Cell Res., 30, 2020
|
|
7EBK
 
 | |
7EBJ
 
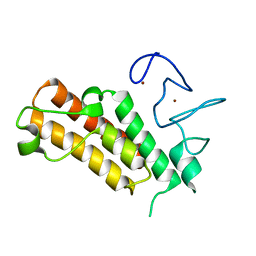 | | Apo structure of the mouse Trim66 PHD-Bromo dual domain | | Descriptor: | Tripartite motif-containing protein 66, ZINC ION | | Authors: | Wang, Z, Jiang, J. | | Deposit date: | 2021-03-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A TRIM66/DAX1/Dux axis suppresses the totipotent 2-cell-like state in murine embryonic stem cells.
Cell Stem Cell, 29, 2022
|
|
7VV9
 
 | |
7VVG
 
 | |
7VVB
 
 | |
7VV8
 
 | |
7EOS
 
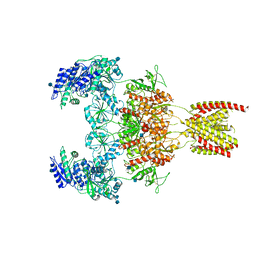 | |
7EOT
 
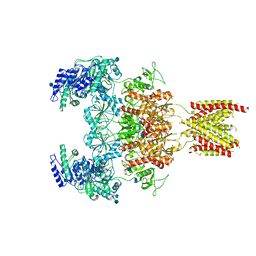 | |
7EOU
 
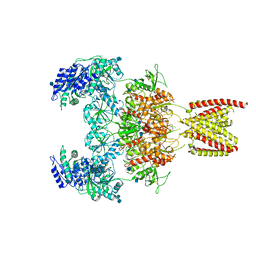 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901/9-AA bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, 9-AMINOACRIDINE, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
7EOQ
 
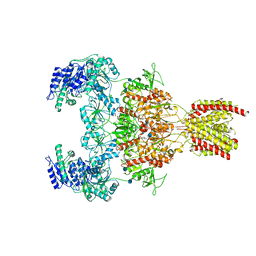 | |
7EOR
 
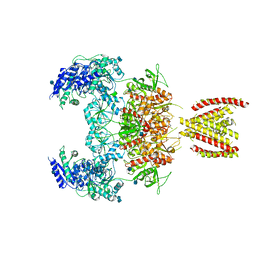 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901 bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, Glutamate receptor ionotropic, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
