6QFQ
 
 | | Structure of human Mcl-1 in complex with indole acid inhibitor | | Descriptor: | 7-(3,5-dimethyl-1~{H}-pyrazol-4-yl)-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QG8
 
 | | Structure of human Bcl-2 in complex with PUMA BH3 peptide | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, Bcl-2-binding component 3 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
1BHR
 
 | | 2'-DEOXY-ISOGUANOSINE BASE PAIRED TO THYMIDINE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*GP*CP*IGUP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Robinson, H, Gao, Y.-G, Bauer, C, Roberts, C, Switzer, C, Wang, A.H.-J. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 2'-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis.
Biochemistry, 37, 1998
|
|
1XZM
 
 | |
1XZH
 
 | |
5JAI
 
 | | Yersinia pestis FabV variant T276G | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
1XZK
 
 | |
5JAM
 
 | | Yersinia pestis FabV variant T276V | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
1XZI
 
 | |
1XZG
 
 | |
5JAQ
 
 | | Yersinia pestis FabV variant T276C | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
7MP4
 
 | | Crystal structure of Epiphyas postvittana antennal carboxylesterase 24 | | Descriptor: | Carboxylesterase-24, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hamiaux, C, Carraher, C. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of an antennally-expressed carboxylesterase suggests lepidopteran odorant degrading enzymes are broadly tuned
Curr Res Insect Sci, 3, 2023
|
|
1XZJ
 
 | |
1XZD
 
 | |
1XZL
 
 | |
1XZF
 
 | |
3CMC
 
 | | Thioacylenzyme intermediate of Bacillus stearothermophilus phosphorylating GAPDH | | Descriptor: | 1,2-ETHANEDIOL, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Moniot, S, Vonrhein, C, Bricogne, G, Didierjean, C, Corbier, C. | | Deposit date: | 2008-03-21 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Trapping of the Thioacylglyceraldehyde-3-phosphate Dehydrogenase Intermediate from Bacillus stearothermophilus: DIRECT EVIDENCE FOR A FLIP-FLOP MECHANISM
J.Biol.Chem., 283, 2008
|
|
8CIZ
 
 | | DNA-polymerase sliding clamp (DnaN) from Escherichia coli in complex with Mycoplanecin A. | | Descriptor: | Beta sliding clamp, Mycoplanecin A | | Authors: | Fu, C, Liu, Y, Walt, C, Bader, C, Rasheed, S, Lukat, P, Neuber, M, Blankenfeldt, W, Kalinina, O, Mueller, R. | | Deposit date: | 2023-02-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Elucidation of unusual biosynthesis and DnaN-targeting mode of action of potent anti-tuberculosis antibiotics Mycoplanecins.
Nat Commun, 15, 2024
|
|
6T5L
 
 | |
8CIX
 
 | | DNA-polymerase sliding clamp (DnaN) from Escherichia coli in complex with Griselimycin. | | Descriptor: | ACETATE ION, Beta sliding clamp, CALCIUM ION, ... | | Authors: | Fu, C, Liu, Y, Walt, C, Bader, C, Rasheed, S, Lukat, P, Neuber, M, Blankenfeldt, W, Kalinina, O, Mueller, R. | | Deposit date: | 2023-02-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidation of unusual biosynthesis and DnaN-targeting mode of action of potent anti-tuberculosis antibiotics Mycoplanecins.
Nat Commun, 15, 2024
|
|
6T7D
 
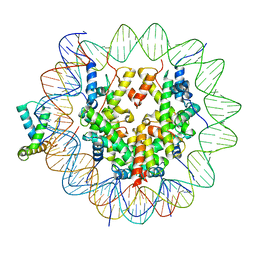 | | Structure of human Sox11 transcription factor in complex with a nucleosome | | Descriptor: | DNA (151-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Dodonova, S.O, Zhu, F, Dienemann, C, Taipale, J, Cramer, P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Nature, 580, 2020
|
|
6TEP
 
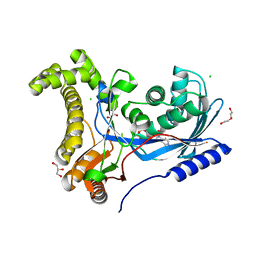 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
6T5V
 
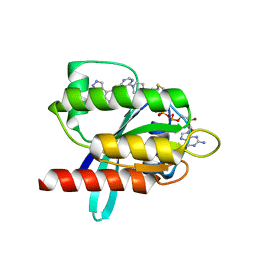 | | KRasG12C ligand complex | | Descriptor: | 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
6TGN
 
 | |
6TCD
 
 | | Crystal structure of Salmo salar RidA-2 | | Descriptor: | ACETATE ION, Ribonuclease UK114, SULFATE ION | | Authors: | Ricagno, S, Visentin, C, Di Pisa, F, Digiovanni, S, Oberti, L, Degani, G, Popolo, L, Bartorelli, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.
Sci Rep, 10, 2020
|
|
