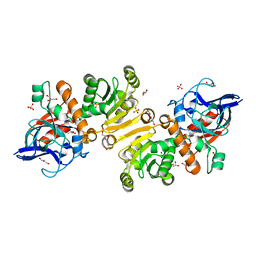
1H0K
 
 | |
7R6R
 
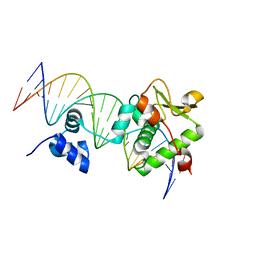 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
1GQ7
 
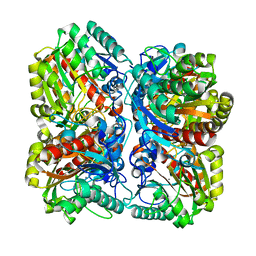 | | PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS | | Descriptor: | MANGANESE (II) ION, PROCLAVAMINATE AMIDINO HYDROLASE | | Authors: | Elkins, J.M, Clifton, I.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Hewitson, K.S. | | Deposit date: | 2001-11-20 | | Release date: | 2002-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Oligomeric structure of proclavaminic acid amidino hydrolase: evolution of a hydrolytic enzyme in clavulanic acid biosynthesis.
Biochem. J., 366, 2002
|
|
1GZE
 
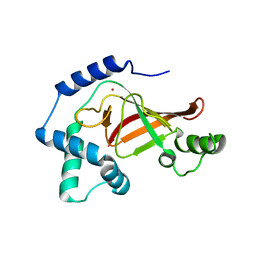 | | Structure of the Clostridium botulinum C3 exoenzyme (L177C mutant) | | Descriptor: | MERCURY (II) ION, MONO-ADP-RIBOSYLTRANSFERASE C3 | | Authors: | Menetrey, J, Flatau, G, Stura, E.A, Charbonnier, J.B, Gas, F, Teulon, J.M, Le Du, M.H, Boquet, P, Menez, A. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nad Binding Induces Conformational Changes in Rho Adp-Ribosylating Clostridium Botulinum C3 Exoenzyme
J.Biol.Chem., 277, 2002
|
|
1GU7
 
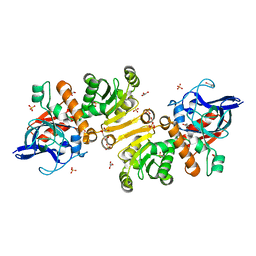 | | Enoyl thioester reductase from Candida tropicalis | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1,MITOCHONDRIAL, GLYCEROL, ... | | Authors: | Airenne, T.T, Torkko, J.M, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1H2K
 
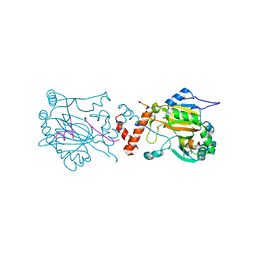 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | FACTOR INHIBITING HIF1, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1GQW
 
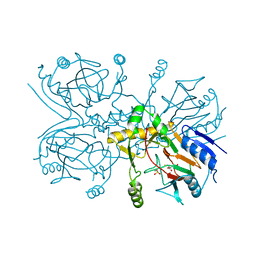 | | Taurine/alpha-ketoglutarate Dioxygenase from Escherichia coli | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ALPHA-KETOGLUTARATE-DEPENDENT TAURINE DIOXYGENASE, ... | | Authors: | Elkins, J.M, Ryle, M.J, Clifton, I.J, Dunning-Hotopp, J.C, Lloyd, J.S, Burzlaff, N.I, Baldwin, J.E, Hausinger, R.P, Roach, P.L. | | Deposit date: | 2001-12-05 | | Release date: | 2002-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Crystal Structure of Escherichia Coli Taurine/Alpha-Ketoglutarate Dioxygenase Complexed to Ferrous Iron and Substrates
Biochemistry, 41, 2002
|
|
1GZF
 
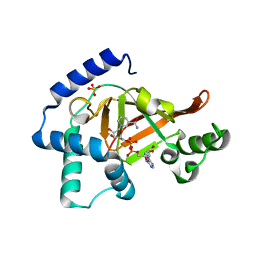 | | Structure of the Clostridium botulinum C3 exoenzyme (wild-type) in complex with NAD | | Descriptor: | 3-(AMINOCARBONYL)-1-[(3R,4S,5R)-3,4-DIHYDROXY-5-METHYLTETRAHYDRO-2-FURANYL]PYRIDINIUM, ADENOSINE-5'-DIPHOSPHATE, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Menetrey, J, Flatau, G, Stura, E.A, Charbonnier, J.B, Gas, F, Teulon, J.M, Le Du, M.H, Boquet, P, Menez, A. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nad Binding Induces Conformational Changes in Rho Adp-Ribosylating Clostridium Botulinum C3 Exoenzyme
J.Biol.Chem., 277, 2002
|
|
1H2M
 
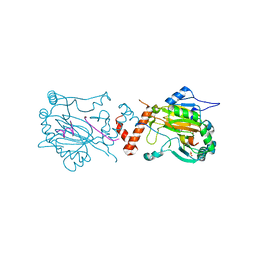 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | FACTOR INHIBITING HIF1, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA, N-OXALYLGLYCINE, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1H2N
 
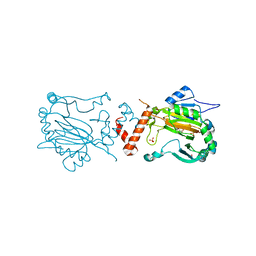 | | Factor Inhibiting HIF-1 alpha | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1H2L
 
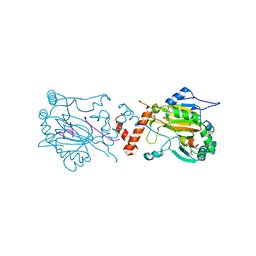 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FACTOR INHIBITING HIF1, FE (II) ION, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1GZ7
 
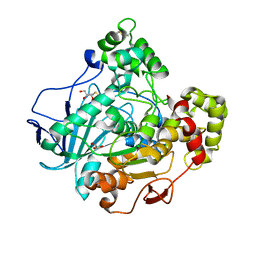 | | Crystal structure of the closed state of lipase 2 from Candida rugosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LIPASE 2 | | Authors: | Mancheno, J.M, Hermoso, J.A. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights Into the Lipase/Esterase Behavior in the Candida Rugosa Lipases Family: Crystal Structure of the Lipase 2 Isoenzyme at 1.97A Resolution
J.Mol.Biol., 332, 2003
|
|
1HA6
 
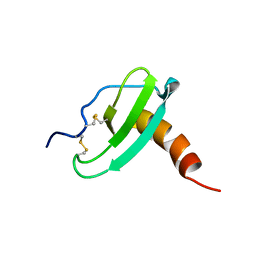 | | NMR Solution Structure of Murine CCL20/MIP-3a Chemokine | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN 3 ALPHA | | Authors: | Perez-Canadillas, J.M, Zaballos, A, Gutierrez, J, Varona, R, Roncal, F, Albar, J.P, Marquez, G, Bruix, M. | | Deposit date: | 2001-03-28 | | Release date: | 2001-08-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Murine Ccl20/Mip3-A, a Chemiokine that Specifically Chemoattracs Immature Dendritic Cells and Lymphocytes Through its Highly Specific Interaction with the Beta-Chemokine Receptor Ccr6
J.Biol.Chem., 276, 2001
|
|
1GYR
 
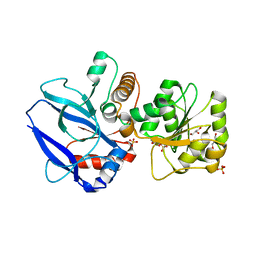 | | Mutant form of enoyl thioester reductase from Candida tropicalis | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, GLYCEROL, SULFATE ION | | Authors: | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-04-29 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1GQ6
 
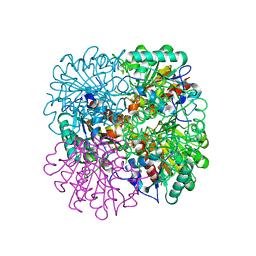 | | PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS | | Descriptor: | MANGANESE (II) ION, PROCLAVAMINATE AMIDINO HYDROLASE | | Authors: | Elkins, J.M, Clifton, I.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Hewitson, K.S. | | Deposit date: | 2001-11-20 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligomeric Structure of Proclavaminic Acid Amidino Hydrolase: Evolution of a Hydrolytic Enzyme in Clavulanic Acid Biosynthesis
Biochem.J., 366, 2002
|
|
1HQ3
 
 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | Descriptor: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | Authors: | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | Deposit date: | 2000-12-14 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HNN
 
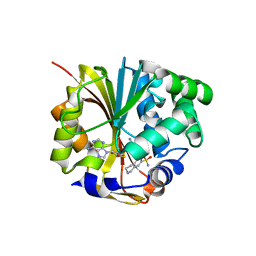 | | CRYSTAL STRUCTURE OF HUMAN PNMT COMPLEXED WITH SK&F 29661 AND ADOHCY(SAH) | | Descriptor: | 1,2,3,4-TETRAHYDRO-ISOQUINOLINE-7-SULFONIC ACID AMIDE, PHENYLETHANOLAMINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Martin, J.L, Begun, J, McLeish, M.J, Caine, J.M, Grunewald, G.L. | | Deposit date: | 2000-12-07 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Getting the adrenaline going: crystal structure of the adrenaline-synthesizing enzyme PNMT.
Structure, 9, 2001
|
|
1HTX
 
 | | SOLUTION STRUCTURE OF THE MAIN ALPHA-AMYLASE INHIBITOR FROM AMARANTH SEEDS | | Descriptor: | ALPHA-AMYLASE INHIBITOR AAI | | Authors: | Martins, J.C, Enassar, M, Willem, R, Wieruzeski, J.M, Lippens, G, Wodak, S.J. | | Deposit date: | 2001-01-02 | | Release date: | 2001-07-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the main alpha-amylase inhibitor from amaranth seeds.
Eur.J.Biochem., 268, 2001
|
|
5ISI
 
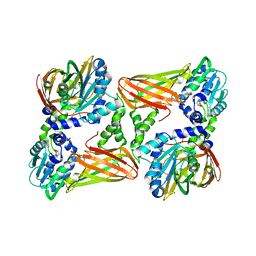 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0920 (5'-amino-5'-deoxyadenosine) | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 5'-amino-5'-deoxyadenosine, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0920 (5'-amino-5'-deoxyadenosine)
To Be Published
|
|
2RK1
 
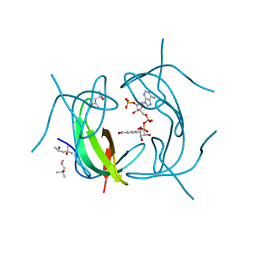 | | DHFR R67 Complexed with NADP and dihydrofolate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DIHYDROFOLIC ACID, Dihydrofolate reductase type 2, ... | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of a type II dihydrofolate reductase catalytic ternary complex.
Biochemistry, 46, 2007
|
|
4LXA
 
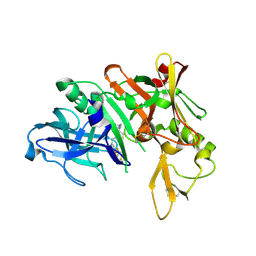 | | Crystal Structure of Human Beta Secretase in Complex with Compound 11a | | Descriptor: | (1R,3S,4S,5R)-3-{4-amino-3-fluoro-5-[(1,1,1,3,3,3-hexafluoropropan-2-yl)oxy]benzyl}-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1-oxide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2013-07-29 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of cyclic sulfoxide hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LXV
 
 | | Crystal Structure of the Hemagglutinin from a H1N1pdm A/WASHINGTON/5/2011 virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Chang, J.C, Guo, Z, Carney, P.J, Shore, D.A, Donis, R.O, Cox, N.J, Villanueva, J.M, Klimov, A.I, Stevens, J. | | Deposit date: | 2013-07-30 | | Release date: | 2014-02-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Stability of Influenza A(H1N1)pdm09 Virus Hemagglutinins.
J.Virol., 88, 2014
|
|
2RH2
 
 | | High Resolution DHFR R-67 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Dihydrofolate reductase type 2 | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal Structure of a Type II Dihydrofolate Reductase Catalytic Ternary Complex
Biochemistry, 46, 2007
|
|
4M0A
 
 | | Human DNA Polymerase Mu post-catalytic complex | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA-directed DNA/RNA polymerase mu, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2013-08-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4M21
 
 | | Crystal Structure of small molecule acrylamide 11 covalently bound to K-Ras G12C | | Descriptor: | 1-(4-{[(4,5-dichloro-2-methoxyphenyl)amino]acetyl}piperazin-1-yl)propan-1-one, GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
