3B3Q
 
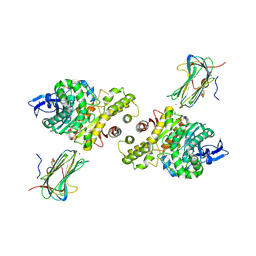 | | Crystal structure of a synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, X, Liu, H, Shim, A, Focia, P, He, X. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5LAE
 
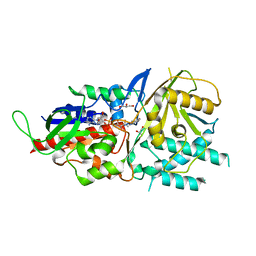 | | Crystal structure of murine N1-acetylpolyamine oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Snijder, A, Barlind, L. | | Deposit date: | 2016-06-14 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
8H9O
 
 | |
8H9W
 
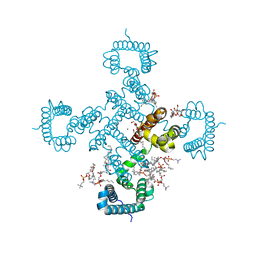 | |
8HA2
 
 | |
8H9X
 
 | |
8H9Y
 
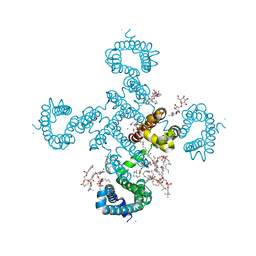 | |
8HA1
 
 | |
8GRF
 
 | | Crystal structure of F-box protein in the ternary complex with adaptor protein Skp1(DL) and its substrate | | Descriptor: | 1,2-ETHANEDIOL, Citrate synthase, E3 ubiquitin ligase complex SCF subunit, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GR9
 
 | | Crystal structure of peroxisomal citrate synthase (Cit2) from Saccharomyces cerevisiae in complex with oxaloacetate and coenzyme-A | | Descriptor: | CHLORIDE ION, COENZYME A, Citrate synthase, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GR8
 
 | | Crystal structure of peroxisomal citrate synthase (Cit2) from Saccharomycescerevisiae | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Citrate synthase, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GRE
 
 | | F-box protein in complex with skp1(FL) and substrate | | Descriptor: | Citrate synthase, E3 ubiquitin ligase complex SCF subunit, F-box protein UCC1, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GQZ
 
 | | Crystal structure of mitochondrial citrate synthase (Cit1) from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Citrate synthase, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8IF4
 
 | | Structure of human alpha-2/delta-1 without mirogabalin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Voltage-dependent calcium channel subunit alpha-2/delta-1 | | Authors: | Kozai, D, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Recognition Mechanism of a Novel Gabapentinoid Drug, Mirogabalin, for Recombinant Human alpha 2 delta 1, a Voltage-Gated Calcium Channel Subunit.
J.Mol.Biol., 435, 2023
|
|
8IF3
 
 | | Structure of human alpha-2/delta-1 with mirogabalin | | Descriptor: | 2-[(1R,5S,6S)-6-(aminomethyl)-3-ethyl-6-bicyclo[3.2.0]hept-3-enyl]acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kozai, D, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Recognition Mechanism of a Novel Gabapentinoid Drug, Mirogabalin, for Recombinant Human alpha 2 delta 1, a Voltage-Gated Calcium Channel Subunit.
J.Mol.Biol., 435, 2023
|
|
6KCS
 
 | | Crystal structure of HIRAN domain of HLTF in complex with duplex DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*AP*CP*GP*TP*AP*CP*AP*GP*T)-3'), Helicase-like transcription factor | | Authors: | Hishiki, A, Hashimoto, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of HIRAN domain of human HLTF bound to duplex DNA provides structural basis for DNA unwinding to initiate replication fork regression.
J.Biochem., 167, 2020
|
|
5LFO
 
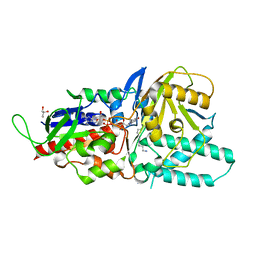 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5LGB
 
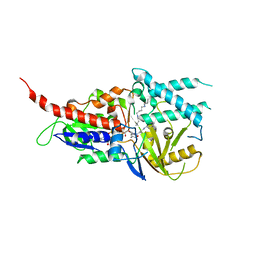 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with MDL72527 | | Descriptor: | FAD-MDL72527 adduct, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-06 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5MBX
 
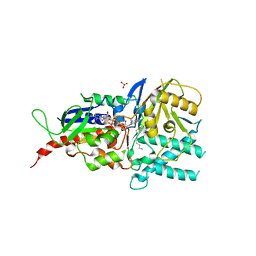 | | Crystal structure of reduced murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5T87
 
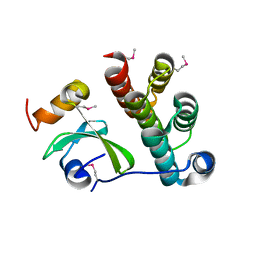 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | Descriptor: | CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
7EVH
 
 | | Odinarchaeota tubulin H393D mutant, in a psuedo protofilament arrangement, bound to 59% GDP, 41% phosphate | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Tubulin-like protein | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVI
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 79% GTP, 21% GDP, Na+, Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7F1A
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound 78% GTP/22% GDP 1 K+, 1 Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-06-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVG
 
 | |
7EVB
 
 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 77% GTP/23% and 2 Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
