5XVF
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH062 | | Descriptor: | 2-(4-azanylpiperidin-1-yl)-6-chloranyl-N-(1-methylimidazol-4-yl)quinazolin-4-amine, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
7XP7
 
 | | Crystal Structure of the Flavoprotein ColB1 Catalyzing Assembly Line-Tethered Cysteine Dehydrogenation | | Descriptor: | Cyclohexanecarboxyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ma, X.Y, Tang, Z.J, Liu, W, Ma, M. | | Deposit date: | 2022-05-03 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Mechanistic Insights into ColB1, a Flavoprotein Functioning in-trans in the 2,2'-Bipyridine Assembly Line for Cysteine Dehydrogenation.
Acs Chem.Biol., 18, 2023
|
|
9JHA
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with BD626-HTL substrate | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]-2-[10,10-dimethyl-3-[(1~{R},5~{S})-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]-6-[(1~{R},5~{S})-3-oxa-8-azoniabicyclo[3.2.1]octan-8-ylidene]anthracen-9-yl]benzoic acid, CHLORIDE ION, ... | | Authors: | Zhixing, C, Kecheng, Z. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A palette of bridged bicycle-strengthened fluorophores.
Nat.Methods, 22, 2025
|
|
5ZNT
 
 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
5ZNS
 
 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
3H16
 
 | | Crystal structure of a bacteria TIR domain, PdTIR from Paracoccus denitrificans | | Descriptor: | SULFATE ION, TIR protein | | Authors: | Chan, S.L, Low, L.Y, Santelli, E, Pascual, J. | | Deposit date: | 2009-04-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mimicry in Innate Immunity: CRYSTAL STRUCTURE OF A BACTERIAL TIR DOMAIN.
J.Biol.Chem., 284, 2009
|
|
7XXU
 
 | | Macaca fascicularis galectin-10/Charcot-Leyden crystal protein | | Descriptor: | Galectin | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8G4M
 
 | | Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Wang, S, Morano, N.C, Shapiro, L, Kwong, P.D. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide.
Cell Rep, 42, 2023
|
|
8G4T
 
 | | Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein BG505 DS-SOSIP gp120, ... | | Authors: | Wang, S, Kwong, P.D. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide.
Cell Rep, 42, 2023
|
|
4PB9
 
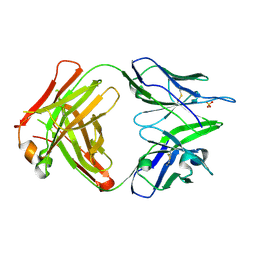 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab64 | | Descriptor: | Ab64 heavy chain, Ab64 light chain, SULFATE ION | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-11 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
4PB0
 
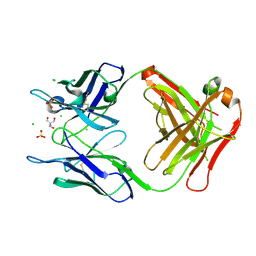 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab53 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ab53 heavy chain, Ab53 light chain, ... | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
7XCT
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XCR
 
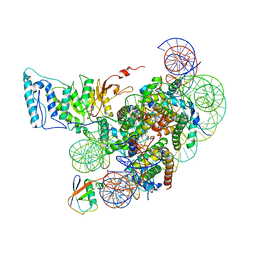 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD0
 
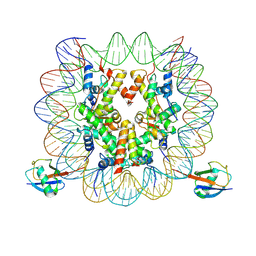 | | cryo-EM structure of H2BK34ub nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD1
 
 | | cryo-EM structure of unmodified nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7YNX
 
 | | Crystal structure of Pirh2 bound to poly-Ala peptide | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of an Ala-rich C-degron by the E3 ligase Pirh2.
Nat Commun, 14, 2023
|
|
7ZRB
 
 | |
7XXY
 
 | | Macaca mulatta galectin-10/Charcot-Leyden crystal protein with lactose | | Descriptor: | Galectin-10/Charcot-Leyden crystal protein, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
7XXX
 
 | | Macaca mulatta galectin-10/Charcot-Leyden crystal protein | | Descriptor: | Galectin-10/Charcot-Leyden crystal protein | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
7XXZ
 
 | | Macaca mulatta galectin-10/Charcot-Leyden crystal protein with glycerol | | Descriptor: | GLYCEROL, Galectin-10/Charcot-Leyden crystal protein | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
7XXW
 
 | |
7XXV
 
 | | Macaca fascicularis galectin-10/Charcot-Leyden crystal protein with lactose | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y. | | Deposit date: | 2022-05-31 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Glutathione disrupts galectin-10 Charcot-Leyden crystal formation to possibly ameliorate eosinophil-based diseases such as asthma.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
7E3O
 
 | |
3LGS
 
 | |
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
