4MWJ
 
 | | Anhui N9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWW
 
 | | Shanghai N9-oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWQ
 
 | | Anhui N9-oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
3TI8
 
 | | Crystal structure of influenza A virus neuraminidase N5 complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
4MWY
 
 | | Shanghai N9-laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
3TI3
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3SJ9
 
 | | crystal structure of the C147A mutant 3C of CVA16 in complex with FAGLRQAVTQ peptide | | Descriptor: | 3C protease, FAGLRQAVTQ peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3SJK
 
 | | Crystal structure of the C147A mutant 3C from enterovirus 71 | | Descriptor: | 3C protease, KPVLRTATVQGPSLDF peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
6LXI
 
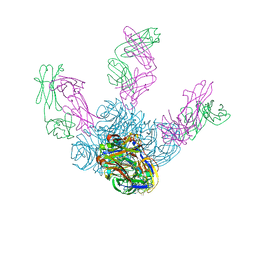 | | Crystal structure of Z2B3 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | CALCIUM ION, Heavy chain of Z2B3 Fab, Light chain of Z2B3 Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
8J5J
 
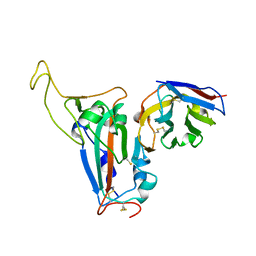 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
8XSI
 
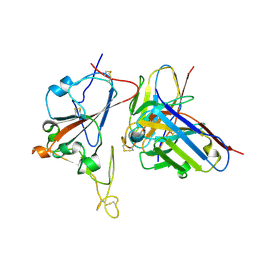 | | SARS-CoV-2 RBD + IMCAS-364 (Local Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-364 H chain, IMCAS-364 L chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSE
 
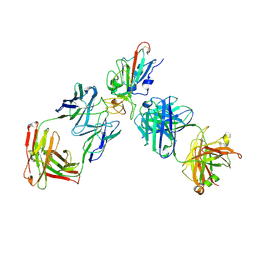 | | SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 H chain, IMCAS-123 L chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSJ
 
 | | SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSL
 
 | | SARS-CoV-2 spike + IMCAS-123 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 heavy chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSF
 
 | | SARS-CoV-2 RBD + IMCAS-364 + hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, IMCAS-364 H chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
3P2T
 
 | | Crystal Structure of Leukocyte Ig-like Receptor LILRB4 (ILT3/LIR-5/CD85k) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 4, SULFATE ION | | Authors: | Chen, Y, Nam, G, Cheng, H, Zhang, J.H, Willcox, B.E, Gao, G.F. | | Deposit date: | 2010-10-04 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): a myeloid inhibitory receptor involved in immune tolerance
J.Biol.Chem., 286, 2011
|
|
3Q2C
 
 | | Binding properties to HLA class I molecules and the structure of the leukocyte Ig-like receptor A3 (LILRA3/ILT6/LIR4/CD85e) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily A member 3 | | Authors: | Ryu, M, Chen, Y, Qi, J.X, Liu, J, Shi, Y, Cheng, H, Gao, G.F. | | Deposit date: | 2010-12-20 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence
Plos One, 6, 2011
|
|
3NSS
 
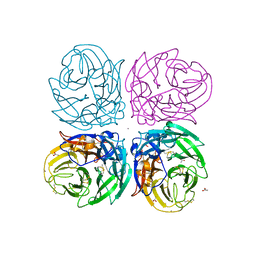 | | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active sites | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J.X, Zhang, W, Vavricka, C.J, Shi, Y, Gao, G.F. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site
Nat.Struct.Mol.Biol., 17, 2010
|
|
4EWJ
 
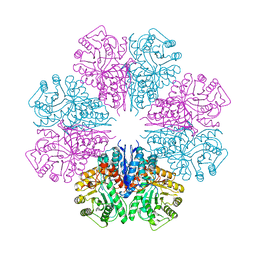 | | structure of the enloase from Streptococcus suis serotype 2 | | Descriptor: | Enolase 2 | | Authors: | Lu, Q, Lu, H, Qi, J, Lu, G, Gao, G.F. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | An octamer of enolase from Streptococcus suis.
Protein Cell, 3, 2012
|
|
4EQ2
 
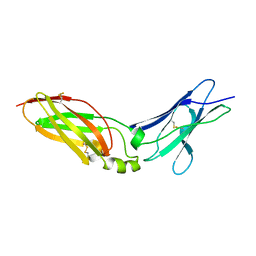 | | Crystal Structure Analysis of Chicken Interferon Gamma Receptor Alpha Chain | | Descriptor: | Interferon gamma receptor 1 | | Authors: | Ping, Z, Qi, J, Lu, G, Shi, Y, Wang, X, Gao, G.F, Wang, M. | | Deposit date: | 2012-04-18 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure of the interferon gamma receptor alpha chain from chicken reveals an undetected extra helix compared with the human counterparts.
J.Interferon Cytokine Res., 34, 2014
|
|
4F7T
 
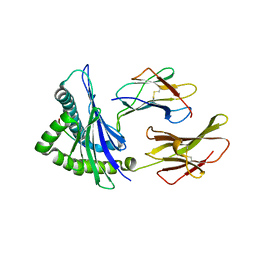 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PB1 (498-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
4F7P
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009H1N1 PB1 (496-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
4EQ3
 
 | | Crystal Structure Analysis of Selenomethionine (Se-Met) Substituted Chicken Interferon Gamma Receptor Alpha Chain | | Descriptor: | Interferon gamma receptor 1 | | Authors: | Ping, Z, Qi, J, Lu, G, Shi, Y, Wang, X, Gao, G.F, Wang, M. | | Deposit date: | 2012-04-18 | | Release date: | 2013-04-24 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of the interferon gamma receptor alpha chain from chicken reveals an undetected extra helix compared with the human counterparts.
J.Interferon Cytokine Res., 34, 2014
|
|
7EMC
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-THR-GLU-ILE-ARG-GLU-LEU-LEU-VAL, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMB
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-ALA-ALA-ILE-GLU-GLU-GLU-ASP-ILE, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
