7V94
 
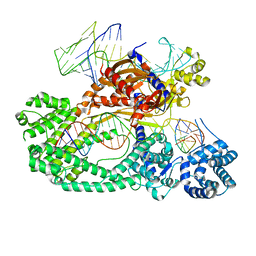 | | Cryo-EM structure of the Cas12c2-sgRNA-target DNA ternary complex | | Descriptor: | Cas12c2, sgRNA, target DNA (non target strand), ... | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
5GIJ
 
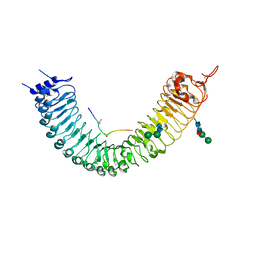 | | Crystal structure of TDR-TDIF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat receptor-like protein kinase TDR, ... | | Authors: | Morita, J, Kato, K, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide
Nat Commun, 7, 2016
|
|
5GLI
 
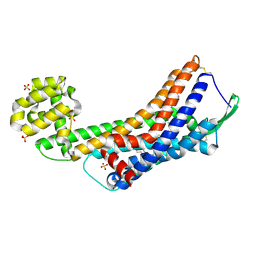 | | Human endothelin receptor type-B in the ligand-free form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Endothelin Receptor Subtype-B, OLEIC ACID, ... | | Authors: | Shihoya, W, Nishizawa, T, Okuta, A, Tani, K, Fujiyoshi, Y, Dohmae, N, Nureki, O, Doi, T. | | Deposit date: | 2016-07-11 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation mechanism of endothelin ETB receptor by endothelin-1.
Nature, 537, 2016
|
|
7V74
 
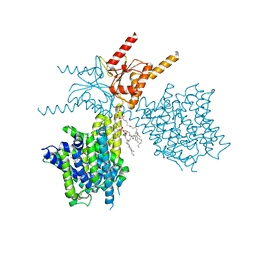 | | Thermostabilized human prestin in complex with sulfate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, SULFATE ION, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
5H35
 
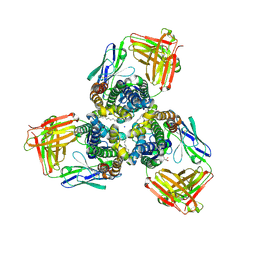 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Sulfolobus solfataricus | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
8XGR
 
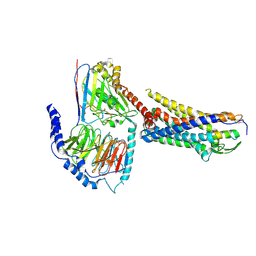 | | ETB-eGt complex bound to endothelin-1 | | Descriptor: | Camelid antibody VHH fragment, Endothelin receptor type B, Endothelin-1, ... | | Authors: | Oshima, H.S, Sano, F.K, Akasaka, H, Iwama, A, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-15 | | Release date: | 2024-04-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Optimizing cryo-EM structural analysis of G i -coupling receptors via engineered G t and Nb35 application.
Biochem.Biophys.Res.Commun., 693, 2024
|
|
8JEW
 
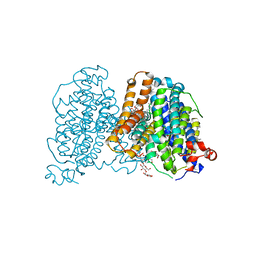 | | Human sodium-dependent vitamin C transporter 1 bound to L-ascorbic acid in an inward-open state | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBIC ACID, ... | | Authors: | Kobayashi, T.A, Kusakizako, T, Nureki, O. | | Deposit date: | 2023-05-16 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Dimeric transport mechanism of human vitamin C transporter SVCT1.
Nat Commun, 15, 2024
|
|
8XWF
 
 | | Structure of CXCR2 bound to CXCL3 (Ligand-receptor focused map) | | Descriptor: | C-X-C chemokine receptor type 2, C-X-C motif chemokine 3 | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XWN
 
 | | Structure of CXCR2 bound to CXCL8 (Ligand-receptor focused map) | | Descriptor: | C-X-C chemokine receptor type 2, MDNCF-a | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XVU
 
 | | Structure of CXCR2 bound to CXCL2 (Ligand-receptor focused map) | | Descriptor: | C-X-C chemokine receptor type 2, C-X-C motif chemokine 2 | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-15 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XX7
 
 | | Structure of CXCR2 bound to CXCL5 (CXCR2-CXCL5-Go Full map) | | Descriptor: | C-X-C chemokine receptor type 2, C-X-C motif chemokine 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XWA
 
 | | Structure of CXCR2 bound to CXCL1 (Ligand-receptor focused map) | | Descriptor: | C-X-C chemokine receptor type 2, Growth-regulated alpha protein | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XWS
 
 | | Structure of CXCR2 bound to CXCL5 (Ligand-receptor focused map) | | Descriptor: | C-X-C chemokine receptor type 2, C-X-C motif chemokine 5 | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XXH
 
 | | Structure of CXCR2 bound to CXCL2 (CXCR2-CXCL2-Go Full map) | | Descriptor: | Antibody fragment ScFv16, C-X-C chemokine receptor type 2, C-X-C motif chemokine 2, ... | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XWM
 
 | | Structure of CXCR2 bound to CXCL6 (Ligand-receptor focused map) | | Descriptor: | C-X-C chemokine receptor type 2, C-X-C motif chemokine 6 | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XXR
 
 | | Structure of CXCR2 bound to CXCL6 (CXCR2-CXCL6-Go Full map) | | Descriptor: | Antibody fragment ScFv16, C-X-C chemokine receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XX3
 
 | | Structure of CXCR2 bound to CXCL3 (CXCR2-CXCL3-Go Full map) | | Descriptor: | Antibody fragment ScFv16, C-X-C chemokine receptor type 2, C-X-C motif chemokine 3, ... | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
7VTI
 
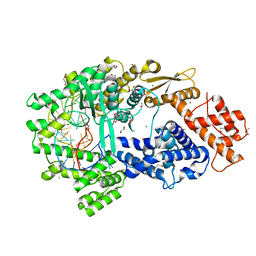 | | Crystal structure of the Cas13bt3-crRNA binary complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Nakagawa, R, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-29 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
8XX6
 
 | | Structure of CXCR2 bound to CXCL8 (CXCR2-CXCL8-Go Full map) | | Descriptor: | Antibody fragment ScFv16, C-X-C chemokine receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XXX
 
 | | Structure of CXCR2 bound to CXCL6 (Composite map) | | Descriptor: | Antibody fragment ScFv16, C-X-C chemokine receptor type 2, C-X-C motif chemokine 6, ... | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
8XWV
 
 | | Structure of CXCR2 bound to CXCL1 (CXCR2-CXCL1-Go Full map) | | Descriptor: | Antibody fragment ScFv16, C-X-C chemokine receptor type 2, Growth-regulated alpha protein, ... | | Authors: | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Mol.Cell, 85, 2025
|
|
7VTN
 
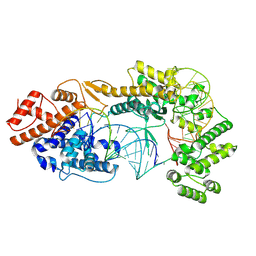 | | Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex | | Descriptor: | Cas13bt3, crRNA, target RNA | | Authors: | Nakagawa, R, Soumya, K, Han, A, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Tomohiro, N, Yamashita, K, Feng, Z, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-30 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
7VVO
 
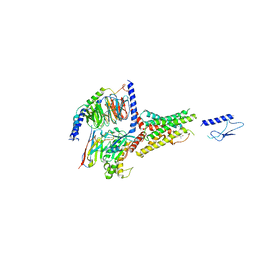 | | PTH-bound human PTH1R in complex with Gs (class5) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVJ
 
 | | PTHrP-bound human PTH1R in complex with Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VR8
 
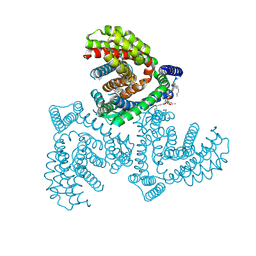 | | Inward-facing structure of human EAAT2 in the substrate-free state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
