8D1F
 
 | | hBest2 5mM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.82 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1E
 
 | | hBest2 1uM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.78 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1L
 
 | | hBest1 Ca2+-bound partially open aperture state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1M
 
 | |
8D1J
 
 | | hBest1 5mM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1H
 
 | | hBest2 Ca2+-unbound closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
7LHZ
 
 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid (compound 25) | | Descriptor: | (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*AP*TP*CP*AP*TP*AP*CP*AP*AP*CP*GP*TP*AP*A)-3'), ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2021-01-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery and Optimization of DNA Gyrase and Topoisomerase IV Inhibitors with Potent Activity against Fluoroquinolone-Resistant Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
5MEV
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[(3,4-dichlorophenyl)methyl]-8-methyl-13-[(4-methylsulfonylphenyl)methyl]-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20,22,24-triene-3,7,15,26-tetrone, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Acs Med.Chem.Lett., 8, 2017
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
1RHM
 
 | |
1RHK
 
 | |
1RHR
 
 | |
1RHJ
 
 | |
1OQF
 
 | | Crystal structure of the 2-methylisocitrate lyase | | Descriptor: | 2-methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-03-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
2HJP
 
 | |
2DUA
 
 | |
2HRW
 
 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
7LRT
 
 | |
7LRS
 
 | |
7P0Z
 
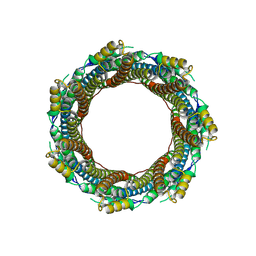 | | 2.43 A Mycobacterium marinum EspB. | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Gijsbers, A, Zhang, Y, Vinciauskaite, V, Siroy, A, Ye, G, Tria, G, Mathew, A, Sanchez-Puig, N, Lopez-Iglesias, C, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Priming mycobacterial ESX-secreted protein B to form a channel-like structure.
Curr Res Struct Biol, 3, 2021
|
|
8BNS
 
 | |
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
7MM0
 
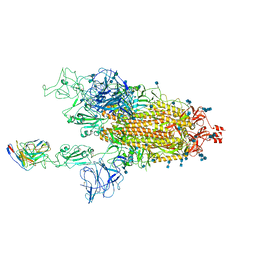 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B1-182.1 Fab heavy chain, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
