5DX5
 
 | | Crystal structure of methionine gamma-lyase from Clostridium sporogenes | | Descriptor: | CHLORIDE ION, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Anufrieva, N.V, Demidkina, T.V. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of methionine gamma-lyase from Clostridium sporogenes.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5ENV
 
 | | YEAST ALCOHOL DEHYDROGENASE WITH BOUND COENZYME | | Descriptor: | Alcohol dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Plapp, B.V, Charlier Jr, H.A, Ramaswamy, S. | | Deposit date: | 2015-11-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic implications from structures of yeast alcohol dehydrogenase complexed with coenzyme and an alcohol.
Arch.Biochem.Biophys., 591, 2015
|
|
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
6EGR
 
 | | Crystal structure of Citrobacter freundii methionine gamma-lyase with V358Y replacement | | Descriptor: | DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Revtovich, S.V, Demitri, N, Raboni, S, Nikulin, A.D, Morozova, E.A, Demidkina, T.V, Storici, P, Mozzarelli, A. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering methionine gamma-lyase from Citrobacter freundii for anticancer activity.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6F3G
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-(5-propan-2-ylpyrrolo[3,2-d]pyrimidin-4-yl)cyclohexane-1,4-diamine | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6F3D
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 4-[4-[[4-(dimethylamino)cyclohexyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]cyclohexane-1-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6F3I
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | (3~{R})-3-[4-[[4-(4-ethanoylpiperazin-1-yl)cyclohexyl]amino]pyrrolo[2,1-f][1,2,4]triazin-5-yl]butanamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6F3E
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 2-[(3~{R})-12-(4-morpholin-4-ylcyclohexyl)oxy-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9,11-tetraen-3-yl]ethanamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
2PMD
 
 | | The structures of aIF2gamma subunit from the archaeon Sulfolobus solfataricus in the GDP-bound form. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PYROPHOSPHATE, ... | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Nikulin, A.D, Hasenohrl, D, Blaesi, U, Manstein, D.J, Fedorov, R.V, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2007-04-21 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | New Insights into the Interactions of the Translation Initiation Factor 2 from Archaea with Guanine Nucleotides and Initiator tRNA.
J.Mol.Biol., 373, 2007
|
|
5ER1
 
 | |
2OAW
 
 | | Structure of SHH variant of "Bergerac" chimera of spectrin SH3 | | Descriptor: | CHLORIDE ION, Spectrin alpha chain, brain | | Authors: | Gabdoulkhakov, A.G, Gushchina, L.V, Nikulin, A.D, Nikonov, S.V, Viguera, A.R, Serrano, L, Filimonov, V.V. | | Deposit date: | 2006-12-18 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic studies of Bergerac-SH3 chimeras.
Biophys.Chem., 139, 2009
|
|
2PLF
 
 | | The structure of aIF2gamma subunit from the archaeon Sulfolobus solfataricus in the nucleotide-free form. | | Descriptor: | Translation initiation factor 2 gamma subunit | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Nikulin, A.D, Hasenohrl, D, Blaesi, U, Manstein, D.J, Fedorov, R.V, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2007-04-19 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New Insights into the Interactions of the Translation Initiation Factor 2 from Archaea with Guanine Nucleotides and Initiator tRNA.
J.Mol.Biol., 373, 2007
|
|
2Q3M
 
 | | Ensemble refinement of the protein crystal structure of an Arabidopsis thaliana putative steroid sulphotransferase | | Descriptor: | Flavonol sulfotransferase-like, MALONIC ACID | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2OHX
 
 | |
5HZB
 
 | | Crystal structure of GII.10 P domain in complex with 2-fucosyllactose (2'FL) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hansman, G.S, Koromyslova, A.D, Singh, B.K.S. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structural Basis for Norovirus Inhibition by Human Milk Oligosaccharides.
J.Virol., 90, 2016
|
|
5HZA
 
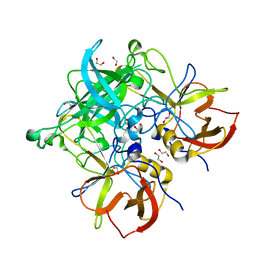 | | Crystal structure of GII.10 P domain in complex with 3-fucosyllactose (3 FL) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]beta-D-glucopyranose | | Authors: | Hansman, G.S, Koromyslova, A.D, Singh, B.K. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis for Norovirus Inhibition by Human Milk Oligosaccharides.
J.Virol., 90, 2016
|
|
3D6G
 
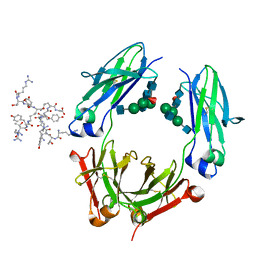 | | Fc fragment of IgG1 (Herceptin) with protein-A mimetic peptide dendrimer ligand. | | Descriptor: | 2-[[(2S)-2,6-bis[[(2S)-2,6-bis[[(2R)-2-[[(2R,3R)-2-[[(2R)-2-amino-5-carbamimidamido-pentanoyl]amino]-3-hydroxy-butanoyl]amino]-3-(4-hydroxyphenyl)propanoyl]amino]hexanoyl]amino]hexanoyl]amino]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Bujacz, A.D, Redzynia, I, Bujacz, G.D, Dinon, F, Pengo, P, Fassina, G. | | Deposit date: | 2008-05-19 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of a Protein A mimetic peptide dendrimer bound to human IgG.
J.Phys.Chem.B, 113, 2009
|
|
3CW2
 
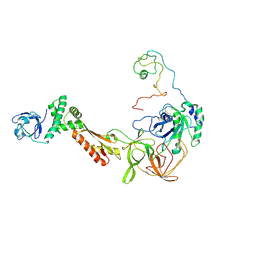 | | Crystal structure of the intact archaeal translation initiation factor 2 from Sulfolobus solfataricus . | | Descriptor: | Translation initiation factor 2 subunit alpha, Translation initiation factor 2 subunit beta, Translation initiation factor 2 subunit gamma | | Authors: | Stolboushkina, E.A, Nikonov, S.V, Nikulin, A.D, Blaesi, U, Manstein, D.J, Fedorov, R.V, Garber, M.B, Nikonov, O.S. | | Deposit date: | 2008-04-21 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the alpha- and beta-subunits.
J.Mol.Biol., 382, 2008
|
|
1WQ1
 
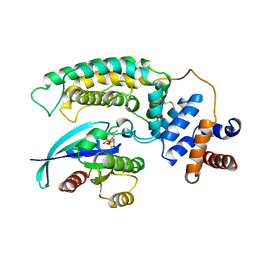 | | RAS-RASGAP COMPLEX | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, H-RAS, ... | | Authors: | Scheffzek, K, Ahmadian, M.R, Kabsch, W, Wiesmueller, L, Lautwein, A, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1997-07-03 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants.
Science, 277, 1997
|
|
1WU1
 
 | |
5MKI
 
 | |
5MKL
 
 | |
3JSY
 
 | | N-terminal fragment of ribosomal protein L10 from Methanococcus jannaschii | | Descriptor: | Acidic ribosomal protein P0 homolog | | Authors: | Kravchenko, O, Mitroshin, I, Nikulin, A.D, Piendl, W, Garber, M, Nikonov, S. | | Deposit date: | 2009-09-11 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a two-domain N-terminal fragment of ribosomal protein L10 from Methanococcus jannaschii reveals a specific piece of the archaeal ribosomal stalk
J.Mol.Biol., 399, 2010
|
|
5N29
 
 | |
5N7M
 
 | |
