2QH1
 
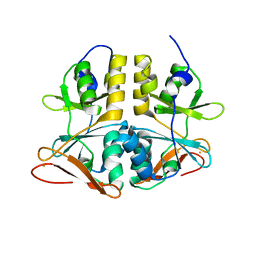 | | Structure of TA289, a CBS-rubredoxin-like protein, in its Fe+2-bound state | | Descriptor: | FE (II) ION, Hypothetical protein Ta0289 | | Authors: | Singer, A.U, Proudfoot, M, Brown, G, Xu, L, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-06-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain.
J.Mol.Biol., 375, 2008
|
|
2QYT
 
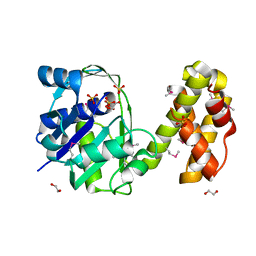 | | Crystal structure of 2-dehydropantoate 2-reductase from Porphyromonas gingivalis W83 | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydropantoate 2-reductase, SULFATE ION | | Authors: | Tan, K, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-15 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of 2-dehydropantoate 2-reductase from Porphyromonas gingivalis W83.
To be Published
|
|
2OT9
 
 | |
2P06
 
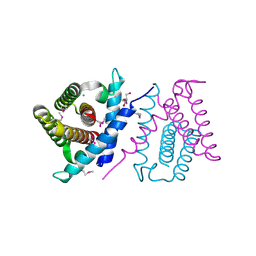 | | Crystal structure of a predicted coding region AF_0060 from Archaeoglobus fulgidus DSM 4304 | | Descriptor: | GLYCEROL, Hypothetical protein AF_0060, MAGNESIUM ION | | Authors: | Nocek, B, Xu, X, Koniyenko, Y, Yakounine, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a predicted coding region AF_0060 from Archaeoglobus fulgidus DSM 4304
To be Published
|
|
2R41
 
 | |
2QLT
 
 | | Crystal structure of an isoform of DL-glycerol-3-phosphatase, Rhr2p, from Saccharomyces cerevisiae | | Descriptor: | (DL)-glycerol-3-phosphatase 1, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of an isoform of DL-glycerol-3-phosphatase, Rhr2p from Saccharomyces cerevisiae.
To be Published
|
|
2RFQ
 
 | | Crystal structure of 3-HSA hydroxylase from Rhodococcus sp. RHA1 | | Descriptor: | 3-HSA hydroxylase, oxygenase, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE | | Authors: | Chang, C, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-01 | | Release date: | 2007-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of 3-HSA hydroxylase, oxygenase from Rhodococcus sp. RHA1.
To be Published
|
|
2PD1
 
 | | Crystal structure of NE2512 protein of unknown function from Nitrosomonas europaea | | Descriptor: | ACETATE ION, Hypothetical protein | | Authors: | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of NE2512 protein of unknown function from Nitrosomonas europaea.
To be Published
|
|
2QMX
 
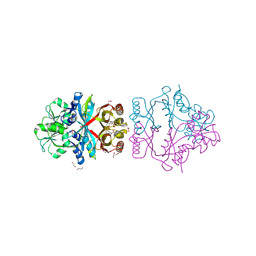 | | The crystal structure of L-Phe inhibited prephenate dehydratase from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHENYLALANINE, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
2QM3
 
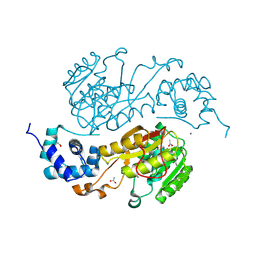 | | Crystal structure of a predicted methyltransferase from Pyrococcus furiosus | | Descriptor: | ACETIC ACID, CALCIUM ION, Predicted methyltransferase | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a predicted methyltransferase from Pyrococcus furiosus.
TO BE PUBLISHED
|
|
2QMM
 
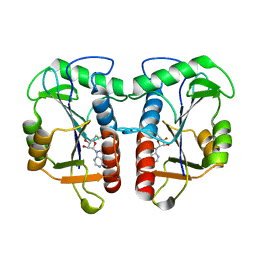 | |
1PQZ
 
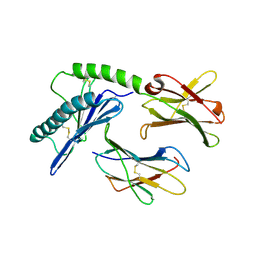 | |
2QNG
 
 | | Crystal structure of unknown function protein SAV2460 | | Descriptor: | CALCIUM ION, Uncharacterized protein SAV2460 | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-18 | | Release date: | 2007-07-31 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of SAV2460.
To be Published
|
|
2QSB
 
 | |
2QNT
 
 | | Crystal structure of protein of unknown function from Agrobacterium tumefaciens str. C58 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Uncharacterized protein Atu1872 | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of protein of unknown function from Agrobacterium tumefaciens str. C58.
To be Published
|
|
2QWV
 
 | | Crystal structure of unknown function protein VCA1059 | | Descriptor: | ACETIC ACID, UPF0217 protein VC_A1059 | | Authors: | Chang, C, Sather, A, Moy, S, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of unknown function protein VCA1059.
To be Published
|
|
2QPV
 
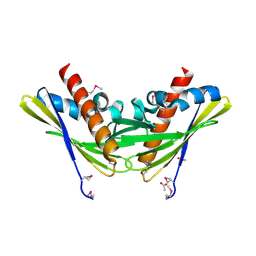 | | Crystal structure of uncharacterized protein Atu1531 | | Descriptor: | ACETIC ACID, Uncharacterized protein Atu1531 | | Authors: | Chang, C, Binkowski, T.A, Xu, X, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-25 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of uncharacterized protein Atu1531.
To be Published
|
|
2FI1
 
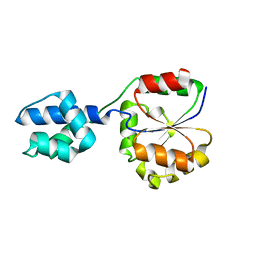 | | The crystal structure of a hydrolase from Streptococcus pneumoniae TIGR4 | | Descriptor: | CALCIUM ION, hydrolase, haloacid dehalogenase-like family | | Authors: | Zhang, R, Zhou, M, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-27 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.4A crystal structure of a hydrolase from Streptococcus pneumoniae TIGR4
To be Published
|
|
2PZ9
 
 | | Crystal structure of putative transcriptional regulator SCO4942 from Streptomyces coelicolor | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Filippova, E.V, Chruszcz, M, Xu, X, Zheng, H, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-17 | | Release date: | 2007-06-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In situ proteolysis for protein crystallization and structure determination.
Nat.Methods, 4, 2007
|
|
1S6Y
 
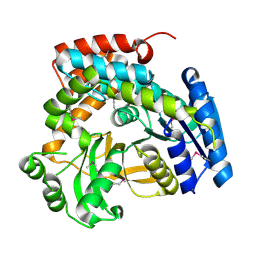 | | 2.3A crystal structure of phospho-beta-glucosidase | | Descriptor: | 6-phospho-beta-glucosidase | | Authors: | Tereshko, V, Dementieva, I, Kim, Y, Collat, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3A CRYSTAL STRUCTURE OF PHOSPHO-BETA-GLUCOSIDASE, licH Gene Product from BACILLUS STEAROTHERMOPHILUS
To be Published
|
|
2QNU
 
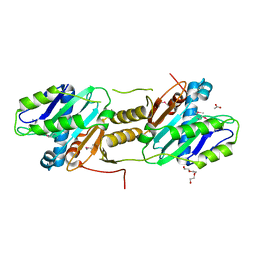 | | Crystal structure of PA0076 from Pseudomonas aeruginosa PAO1 at 2.05 A resolution | | Descriptor: | ACETATE ION, TRIETHYLENE GLYCOL, Uncharacterized protein PA0076 | | Authors: | Filippova, E.V, Chruszcz, M, Skarina, T, Kagan, O, Cymborowski, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Pa0076 from Pseudomonas aeruginosa PAO1 at 2.05 A resolution.
To be Published
|
|
2NPN
 
 | | Crystal structure of putative cobalamin synthesis related protein (CobF) from Corynebacterium diphtheriae | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative cobalamin synthesis related protein, ... | | Authors: | Nocek, B, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of cobalamin synthesis related protein (CobF) from Corynebacterium diphtheriae
To be Published
|
|
2R5R
 
 | | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718 | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, UPF0343 protein NE1163 | | Authors: | Tan, K, Wu, R, Nocek, B, Bigelow, L, Patterson, S, Freeman, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718.
To be Published
|
|
2RBC
 
 | | Crystal structure of a putative ribokinase from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-11-13 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a putative ribokinase from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
1S3J
 
 | | X-ray crystal structure of YusO protein from Bacillus subtilis | | Descriptor: | YusO protein | | Authors: | Osipiuk, J, Wu, R, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-13 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystal structure of YusO protein from Bacillus subtilis, a member of MarR transcriptional regulator family
To be Published
|
|
